Abstract
An erratum was issued for: Chromosome Screening of Human Preimplantation Embryos by Using Spent Culture Medium: Sample Collection and Chromosomal Ploidy Analysis. The Protocol and Representaive Results sections were updated.
In the Protocol, step 3.8.2 was updated from:
After logging into the system, click Create Submission under the NICS tab. Then, select the sequencing platform, choose ChromInst for the reagent, enter the project information in the box under Project ID, set the analysis preferences and upload the files. Once all sequencing files are successfully uploaded, click Submit to start the analysis (Figure 3A).
to:
After logging into the system, click Create Submission under the NICS-A tab. Then, choose NGS for the platform, select corporation, choose ChromInst for the reagent, enter the project information in the box under Project ID, set the analysis preferences and upload the files. Once all sequencing files are successfully uploaded, click Submit to start the analysis (Figure 3A).
In the Representative Results, Figure 3 was updated from:
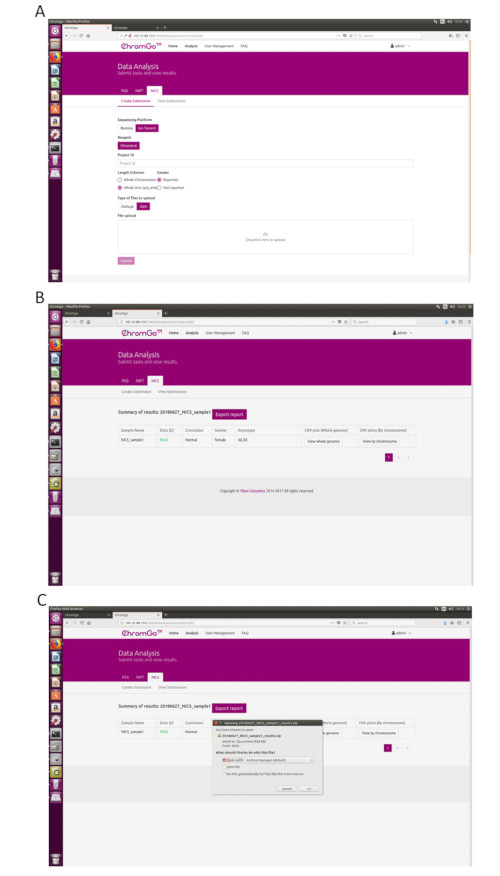
Figure 3. Data Analysis. (A) The page of Create Submission. There are different options for the user application. For sequencing platform, users can choose Illumina or Ion Torrent. For analysis criterion, there are two length detection resolution for selection, the whole chromosome and whole arm level. The users also can choose whether the mosaicism or gender information is reported. Finished the above parameter setting,click on the box under File upload and choose the appropriate sequencing files to upload. For Illumina, choose the files with an extension of fastq.gz. For Ion Torrent platform, choose files with an extension of bam. Click Submit to start the analysis after successfully upload. (B) The view of summary table. The summary table consists of following information: Sample Name: The name of each NICS sample is listed; Data QC: Indicates whether the sequencing file passes the QC for NICS analysis; Conclusion: Indicates whether the NICS analysis is normal or abnormal, "N/A" indicates no conclusive result is available; Gender: If the user chooses to report the sex information, this column will appear in the summary table; Karyotype: Shows the analysis results; CNV plot (Whole Genome): View the CNV profiles of all chromosomes; CNV plot (By Chromosome): View the CNV profiles of each chromosome. (C) The Save Report Page. Click Export report button next to the Summary of Results. Select the information you want to show on the final report and click Export. Select Save File in the appearing dialog window and then click OK. The reports will be saved to the Download folder of the computer. Please click here to view a larger version of this figure.
to:
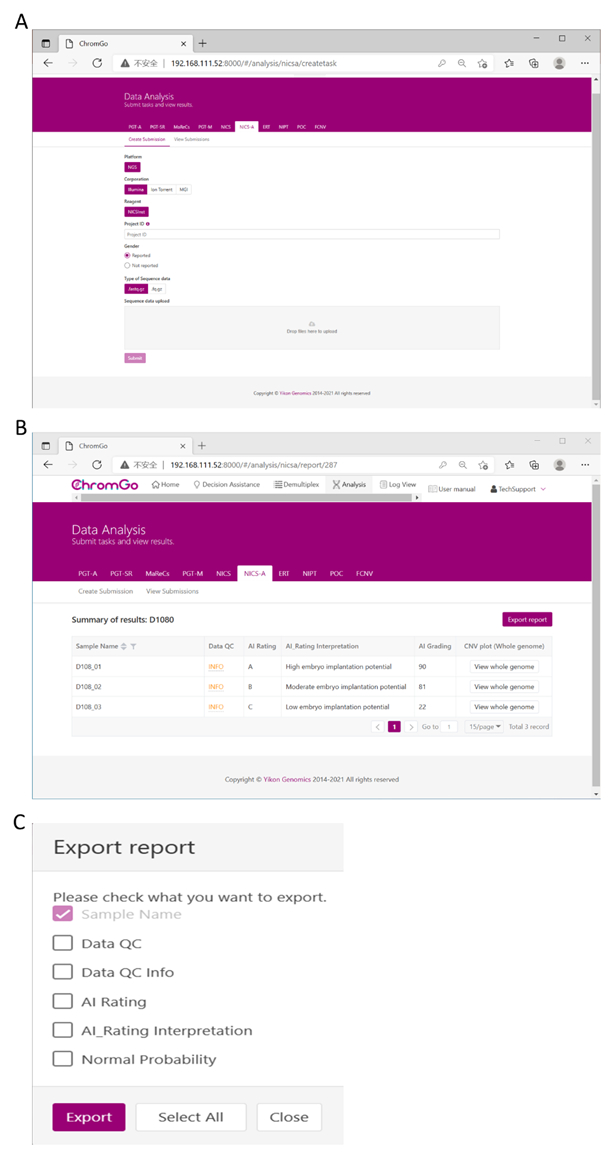
Figure 3. Data Analysis. (A) There are different options for the user application. For sequencing platform corporation, users can choose Illumina, Ion Torrent or MGI. The users can choose whether the gender information is reported. Finished the above parameter setting, click on the box under File upload and choose the appropriate sequencing files to upload. For Illumina, choose the files with an extension of fastq.gz. Click Submit to start the analysis after successfully upload. (B) The view of summary table. The summary table consists of following information: Sample Name: The name of each NICS sample is listed; Data QC: Indicates whether the sequencing file passes the QC for NICS analysis; AI Rating: The rating (A, B or C) for each NICS sample; AI_Rating Interpretation: Evaluation of embryo implantation potential; AI Grading: The score for each NICS sample; CNV plot (Whole Genome): View the CNV profiles of all chromosomes; (C) The Save Report Page. Click Export report button next to the Summary of Results. Select the information you want to show on the final report and click Export. The reports will be saved to the Download folder of your computer. Please click here to view a larger version of this figure.
Protocol
An erratum was issued for: Chromosome Screening of Human Preimplantation Embryos by Using Spent Culture Medium: Sample Collection and Chromosomal Ploidy Analysis. The Protocol and Representaive Results sections were updated.
In the Protocol, step 3.8.2 was updated from:
After logging into the system, click Create Submission under the NICS tab. Then, select the sequencing platform, choose ChromInst for the reagent, enter the project information in the box under Project ID, set the analysis preferences and upload the files. Once all sequencing files are successfully uploaded, click Submit to start the analysis (Figure 3A).
to:
After logging into the system, click Create Submission under the NICS-A tab. Then, choose NGS for the platform, select corporation, choose ChromInst for the reagent, enter the project information in the box under Project ID, set the analysis preferences and upload the files. Once all sequencing files are successfully uploaded, click Submit to start the analysis (Figure 3A).
In the Representative Results, Figure 3 was updated from:

Figure 3. Data Analysis. (A) The page of Create Submission. There are different options for the user application. For sequencing platform, users can choose Illumina or Ion Torrent. For analysis criterion, there are two length detection resolution for selection, the whole chromosome and whole arm level. The users also can choose whether the mosaicism or gender information is reported. Finished the above parameter setting,click on the box under File upload and choose the appropriate sequencing files to upload. For Illumina, choose the files with an extension of fastq.gz. For Ion Torrent platform, choose files with an extension of bam. Click Submit to start the analysis after successfully upload. (B) The view of summary table. The summary table consists of following information: Sample Name: The name of each NICS sample is listed; Data QC: Indicates whether the sequencing file passes the QC for NICS analysis; Conclusion: Indicates whether the NICS analysis is normal or abnormal, "N/A" indicates no conclusive result is available; Gender: If the user chooses to report the sex information, this column will appear in the summary table; Karyotype: Shows the analysis results; CNV plot (Whole Genome): View the CNV profiles of all chromosomes; CNV plot (By Chromosome): View the CNV profiles of each chromosome. (C) The Save Report Page. Click Export report button next to the Summary of Results. Select the information you want to show on the final report and click Export. Select Save File in the appearing dialog window and then click OK. The reports will be saved to the Download folder of the computer. Please click here to view a larger version of this figure.
to:

Figure 3. Data Analysis. (A) There are different options for the user application. For sequencing platform corporation, users can choose Illumina, Ion Torrent or MGI. The users can choose whether the gender information is reported. Finished the above parameter setting, click on the box under File upload and choose the appropriate sequencing files to upload. For Illumina, choose the files with an extension of fastq.gz. Click Submit to start the analysis after successfully upload. (B) The view of summary table. The summary table consists of following information: Sample Name: The name of each NICS sample is listed; Data QC: Indicates whether the sequencing file passes the QC for NICS analysis; AI Rating: The rating (A, B or C) for each NICS sample; AI_Rating Interpretation: Evaluation of embryo implantation potential; AI Grading: The score for each NICS sample; CNV plot (Whole Genome): View the CNV profiles of all chromosomes; (C) The Save Report Page. Click Export report button next to the Summary of Results. Select the information you want to show on the final report and click Export. The reports will be saved to the Download folder of your computer. Please click here to view a larger version of this figure.
Disclosures
No conflicts of interest declared.


