Summary
A method to measure the persistence length or flexural rigidity of biopolymers is described. The method uses a kinesin-driven microtubule gliding assay to experimentally determine the persistence length of individual microtubules and is adaptable to actin-based gliding assays.
Abstract
Microtubules are cytoskeletal polymers which play a role in cell division, cell mechanics, and intracellular transport. Each of these functions requires microtubules that are stiff and straight enough to span a significant fraction of the cell diameter. As a result, the microtubule persistence length, a measure of stiffness, has been actively studied for the past two decades1. Nonetheless, open questions remain: short microtubules are 10-50 times less stiff than long microtubules2-4, and even long microtubules have measured persistence lengths which vary by an order of magnitude5-9.
Here, we present a method to measure microtubule persistence length. The method is based on a kinesin-driven microtubule gliding assay10. By combining sparse fluorescent labeling of individual microtubules with single particle tracking of individual fluorophores attached to the microtubule, the gliding trajectories of single microtubules are tracked with nanometer-level precision. The persistence length of the trajectories is the same as the persistence length of the microtubule under the conditions used11. An automated tracking routine is used to create microtubule trajectories from fluorophores attached to individual microtubules, and the persistence length of this trajectory is calculated using routines written in IDL.
This technique is rapidly implementable, and capable of measuring the persistence length of 100 microtubules in one day of experimentation. The method can be extended to measure persistence length under a variety of conditions, including persistence length as a function of length along microtubules. Moreover, the analysis routines used can be extended to myosin-based acting gliding assays, to measure the persistence length of actin filaments as well.
Introduction
The cytoskeleton, a network of biopolymers found in most eukaryotic cells, plays a role in cellular organization, intracellular transport, and cell mechanics. The mechanical characteristics of the biopolymers of the cytoskeleton (primarily actin and microtubules) play a significant role in determining the mechanical characteristics of the cell as a whole12. Since whole cell mechanics can characterize healthy and diseased cells13,14 and is involved in cellular motility15, the mechanical properties of the underlying cytoskeletal components have been an active area of study for the past two decades1.
The flexibility (or stiffness) of biopolymers is characterized by the persistence length, the length of polymer which bends by approximately one radian under thermal fluctuations at ambient temperature. A number of techniques have been developed to measure persistence length16, for example active techniques which involve bending the polymer using hydrodynamic flow, optical traps, or electric fields4,17,18 , and passive techniques which measure the fluctuations of free polymers in solution5,6 . The active measurements, however, require specialized setups to implement known forces on the micrometer scale, and the free-fluctuation measurements can be challenging due to diffusion out of the plane of focus of the microscope used.
In this article, we describe a complementary, passive, technique to measure the persistence length of microtubules, a cytoskeletal polymer. The technique involves gliding assays, which ensure that the polymer always remains in the focal plane19. Moreover, it involves tracking single fluorophores attached permanently to the polymer of interest, so that specific locations along the polymer are well characterized.
A cartoon of the method is shown in Figure 1. Kinesin moves specifically toward the + end of microtubules, so the microtubules in a gliding assay are propelled unidirectionally. The leading end of the microtubule, beyond the last kinesin attached, is free to fluctuate under the thermal forces of the surrounding solution. As the microtubule is propelled forward, the end fluctuates until binding to a new kinesin molecule further along the glass slide freezes in a given fluctuation. Because kinesin attaches microtubules very strongly, the microtubule is constrained to follow the path of the leading end. Therefore, the statistical fluctuations frozen into the microtubule trajectory are the same as the statistical fluctuations of the free end of microtubules11, and can therefore be used to calculate the persistence length according to20
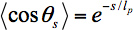
where lp is the persistence length of the microtubule, θs is the angle between tangents to the trajectory separated by a contour length s, and <> denotes an average over all pairs of positions separated by a contour length s.
The gliding assay itself uses kinesin biotinylated at the coiled-coil21 specifically bound to the glass slide via a streptavidin-biotin linkage. This attachment ensures that the motor domains are free to bind to and propel microtubules. In order to follow microtubule trajectories, microtubules are sparsely labeled with organic fluorophores22,23 - the labels must be sparse enough that single fluorophores are resolvable using single molecule fluorescence microscopy. Single fluorophores are tracked using image analysis routines written in IDL. The trajectories of each fluorophore bound to a given microtubule are combined into a composite microtubule trajectory automatically24. The tangent angles θ to each point along a trajectory are calculated; from these tangent angles the <cosθs> value is calculated for each contour length s. Finally, these data are fit to Eq. 1 in order to extract a persistence length for a given microtubule, or for many microtubules in the same gliding assay.
The method is robust enough to work with microtubules prepared in a wide variety of conditions (with different stabilizing agents or other small molecules bound to the microtubule, with bound microtubule associated proteins (MAPs), or with a variety of viscous solutions). In our lab, the technique has been used to characterize the persistence length of microtubules as a function of length along the microtubules and microtubules with different stabilizing agents. The main restriction is that the microtubules must still support kinesin motility. Since kinesin is a robust motor enzyme, this is a fairly loose restriction. By replacing microtubules with actin and kinesin with a myosin family enzyme, the persistence length of actin can be measured using the same technique.
Subscription Required. Please recommend JoVE to your librarian.
Protocol
1. Microtubule Gliding Assay Stock Solutions
Prepare ahead of gliding assay.
- Polymerize 0.5 mg microtubules sparsely labeled with bright organic fluorophore 22. The target label concentration is 1 fluorophore per micrometer of microtubule, or a labeling density of approximately 1 fluorophore per 1,500 tubulin dimers. Store at room temperature, light protected with aluminum, foil for up to two weeks.
- Purify biotin-kinesin21 at approximately 1 μM. Store at -80 °C in 5 μl aliquots for use in individual experiments.
- Prepare the assay buffer (AB) of 50 mM imidiazole, 50 mM potassium chloride (KCl), 4 mM magnesium chloride (MgCl2), 2 mM ethylene glycol tetraacetic acid (EGTA), pH 6.7. Sterile filter and store at 4 °C.
- Dissolve biotinylated bovine serum albumin (biotin-BSA) to 2 mg/ml in AB. Filter with 0.2 μm syringe filter. Store at -80 °C in 100 μl aliquots for long periods, at 4 °C for up to one month.
- Dissolve streptavidin (SA) to 10 mg/ml in AB. Filter with 0.2 μm syringe filter. Store at -80 °C in 20 μl aliquots for long periods, at 4 °C for up to two weeks.
- Dissolve α-casein to 5 mg/ml in AB. Filter with 0.2 μm syringe filter. Store at -80 °C in 100 μl aliquots for long periods, at 4 °C for up to two weeks.
- Dissolve dithiothreitol (DTT) in deionized water at 200 mM. Store at -20 °C in 100 μl aliquots, use within 8 hr of thawing. Can be refrozen.
- Dissolve paclitaxel (PT) in HPLC-grade dimethyl sulfoxide (DMSO) at 4 mM. Store in 10 μl aliquots at -80 °C long term, -20 °C short-term. Use within 8 hours of thawing. Can be refrozen.
- Dissolve glucose in deionized water at 120 mg/ml. Store in 100 μl aliquots at -20 °C. Can be refrozen.
- Dissolve adenosine triphosphate (ATP) in deionized water at 150 mM, pH 7.0. Store in 5 μl aliquots at -80 °C, use within 8 hr of thawing.
- Prepare 100x oxygen scavenging stock25 by dissolving 10,000 units glucose oxidase and 156,000 units catalase into 600 μl total assay buffer. Centrifuge briefly in a microcentrifuge to pellet solids; filter supernatant with 0.2 μm syringe filter. Store at -80 °C in 10 μl aliquots for long periods, at 4 °C for up to one week.
2. Microtubule Gliding Assay, Same Day Solution Preparation
Prepare the solutions in 2.1-2.6 on the day of the experiment. With practice, solutions 2.2-2.6 can be prepared during flow-cell washing. Unless otherwise noted, keep stock solutions on ice.
- Prepare AB, 1 ml of assay buffer with 10 μl of DTT stock (AB with 2 mM DTT). Keep at room temperature.
- Prepare bio-BSA buffer, 40 μl of a 1:1 mixture of the biotin-BSA stock and AB. Keep at room temperature.
- Prepare BSA buffer, mixing 645 μl of AB with 5.5 μl BSA (1 mg/ml BSA in AB). Keep at room temperature.
- Prepare SA buffer, mixing 57 μl of AB with 3 μl streptavidin stock (0.5 mg/ml streptavidin in AB). Keep at room temperature.
- Prepare α-casein buffer, mixing 200 μl of BSA buffer, 50 μl casein stock, and 0.8 μl of 15 μM ATP (1 mg/ml α-casein, 0.8 mg/ml BSA, 50 nM ATP in AB). Keep at room temperature.
- Prepare fluorescence anti-bleach buffer, mixing 95 μl α-casein buffer, 2.5 μl glucose stock, 1 μl 100x oxygen scavenging mix, 1 μl paclitaxel stock, 1 μl 2-mercaptoethanol (βME). Add βME under fume hood. Use fluorescence anti-bleach buffer within 1 hr of preparation. CAUTION: βME is highly toxic and smells awful. Be sure to only open under the fume hood. Store bottle in the absence of light; light degrades βME and its ability to reduce photobleaching.
3. Microtubule Gliding Assay
- Construct about four flow lanes between a 24 x 60 mm coverslip and 22 x 22 mm coverslip using vacuum grease, extruded from a syringe through a pipette tip, as a spacer. Each flow lane should be approximately 10 μl in volume (scale subsequent volumes accordingly).
- Wash 10 μl of bio-BSA buffer into each lane. Incubate 5 min to allow BSA to coat the glass surfaces.
- Wash each lane three times with 15 μl BSA buffer to remove free biotin-BSA, while continuing to block the slide surface. Use a Kimwipe or filter paper to remove buffer from opposite side of flow chamber, being sure not to allow air bubbles to go through the flow chamber.
- Wash 15 μl SA-buffer into each lane. Wait 10 min, or up to 2 hr. The streptavidin will bind to the surface-bound biotin-BSA.
- Wash each lane three times with 15 μl BSA buffer to remove free SA while continuing to block the slide surface.
- Wash each lane with 15 μl α-casein buffer. α-casein helps further block non-specific binding of kinesin and microtubules to the glass.
- Dilute kinesin to 10 nM in α-casein buffer and wash each lane with 15 μl of this kinesin - α-casein solution. Wait 15 min (or up to an hour) for the biotinylated kinesin to bind specifically to the surface-bound streptavidin. The kinesin motor domains will remain free to bind microtubules.
- Dilute paclitaxel to 40 μM in room-temperature α-casein buffer, and wash each lane with 15 μl of this solution to wash out free kinesin and to pre-load each flow chamber with a paclitaxel solution to prevent microtubules from depolymerizing. Cold also depolymerizes microtubules, so make sure these steps occur at room temperature with room temperature solutions.
- Dilute fluorescently labeled microtubules 1:100-1:1,000 in fluorescence anti-bleach buffer with 1 mM ATP. Wash 15 μl into one lane and observe within 30 min.
4. Data Collection
- Observe the microtubules gliding using fluorescence microscopy; a microscope capable of resolving single fluorophores such as a commercial or home-build TIRF setup is required26 (Figure 2).
The microtubules should be propelled by the kinesin motors over the substrate at approximately 0.5 μm/s, depending on temperature. If the field-of-view of the microscope is 50 μm, individual microtubules should be visible for approximately 100 sec. Set illumination intensity so that single fluorophores do not photobleach more quickly than 100 sec. If using laser excitation, a power setting of approximately 3-5 mW is appropriate. - Collect sequences of images for analysis. 600 images at 5 Hz (2 min total) works well. Use sequences long enough that microtubules traverse the entire field-of-view.
5. Data Analysis
An IDL routine, get_lp.pro, is attached. This routine returns a persistence length value based on all microtubules gliding in a given image sequence. Either run this routine on each image sequence, modifying intensity parameters depending on your particular microscope setup, or do the following:
- Track each fluorophore attached to a microtubule to create fluorophore trajectories.
- Combine all the trajectories of fluorophores on a given microtubule into an overall microtubule trajectory; repeat for each microtubule in the image sequence (Figure 3).
- Calculate the tangent angle to each point along the trajectory (Figure 4), and calculate <cosθs> in Eq. 1 by averaging the cosine of the angle difference for every pair of points separated by a path-length s (Figure 5). Find the persistence length for a single microtubule or group of microtubules by fitting <cosθs> to Eq. 1, weighting the individual data points in the fit by the number of independent values of cosθs that are used to compute the average.
Subscription Required. Please recommend JoVE to your librarian.
Representative Results
A snapshot from a gliding assay is shown in Figure 2. A good microtubule density is 1-10 microtubules per field of view; substantially more will result in mistracking as microtubules cross each other. A plot of the 11 microtubule trajectories from the gliding assay in Figure 2 is shown in Figure 3. Typical trajectories are 10 to 30 μm long; some trajectories have gaps where one microtubule crosses another. These trajectories may be discarded from analysis.
A single, long microtubule trajectory is shown in Figure 4A with example tangent angles at two positions along the trajectory. The difference between tangent angles separated by a fixed distance is related to the persistence length; the tangent angle as a function of position along the microtubule trajectory is shown in Figure 4B.
The tangent angle data from many microtubule trajectories is combined to calculate a single persistence length for all microtubules in a given experiment. Figure 5 shows a plot of <cosθs> versus contour length s for the gliding assay of Figure 2. At large contour length values, very few values of cosθ are measured; hence the average is highly variable. The weighted fit conforms to the short s, high-precision data more closely than the long s, low-precision data.
The value of persistence length from these 11 trajectories, 500 ± 40 μm (± standard error of the mean), is representative of persistence lengths for relatively short microtubules2. Similar experiments give a range of persistence lengths between 300 and 1,000 μm.
Troubleshooting
If microtubules are completely absent, increase the concentration of microtubules in step 3.9 to 0.5 mg/ml. If microtubules are still missing, re-polymerize microtubules. Make sure paclitaxel is present in each solution microtubules are diluted into, otherwise microtubules will depolymerize.
If microtubules are freely diffusing in solution but not binding to surface, increase the kinesin concentration in step 3.7 and use AMP-PNP instead of ATP to ensure kinesin binds microtubules irreversibly. If microtubules still do not bind, make new streptavidin stock and buffer, followed by new biotin-BSA buffer. Finally, purify new biotinylated kinesin.
If microtubules bind but do not move, replace ATP stock solution with fresh ATP.
If fluorophores photobleach too quickly, reduce illumination intensity. If fluorophores still photobleach too quickly, replace oxygen scavenging system with fresh stock.
If fluorophores are too dim, increase illumination intensity.

Figure 1. Cartoon of the microtubule gliding assay. Kinesin enzymes are specifically bound to a coverslip by a biotin-streptavidin linkage. Microtubules are sparsely labeled with organic fluorophores. Upon addition of ATP, microtubules are pushed by the kinesin motors. The leading free end of the microtubule fluctuates due to thermal forces in solution; these fluctuations are used to calculate the microtubule persistence length. The length-scale of the microtubule probed by these experiments is the length of the free end.

Figure 2. Typical microscope snapshot of a gliding assay, taken via TIRF microscopy. Microtubules are sparsely decorated with single fluorophores. Scale bar is 5 μm.

Figure 3. Microtubule trajectories from the image sequence shown in Figure 2. Each microtubule trajectory combines many single fluorophore trajectories (about 10 on average), and have been thinned to one point per 100 nm.

Figure 4. Calculating the tangent angles to a trajectory. (A) Trajectory of one microtubule, with example tangent angles (θ) shown. (B) Tangent angle as a function of position along the microtubule trajectory. These data are used to calculate the average angles used in Eq. 1.

Figure 5. Calculating persistence length from tangent angles. (dots) Plot of <cosθs>versus contour length s for the 11 trajectories shown in Figure 3. (solid line) Fit to Eq. 1. For this group of microtubules, the persistence length is 500 ± 40 μm. For long contour lengths (above 10 μm or so), the data are highly variable due to limited statistics.
Subscription Required. Please recommend JoVE to your librarian.
Discussion
Persistence length measurements are a good characterization of the mechanical properties of individual biopolymers. In this article, we have described a method of measuring the persistence length of microtubules. As noted in the introduction, this method is readily extended to examining microtubule mechanical properties in a variety of conditions simply by varying the reagents, temperature, or viscosity in the final step of the gliding assay, 3.9, or by polymerizing microtubules, step 1.1, under different conditions.
The technique itself is quick to perform. Once stock solutions have been prepared ahead of time (step 1), a gliding assay experiment can be performed in 2-3 hr, including up to four variations in conditions on a single microscope slide (the four lanes in step 3.1). Many microtubules can be examined in each experiment; typical experiments in our lab examine 100 microtubules per condition (about 10 image sequences per lane, with 10 microtubules per sequence). The analysis of large data sets is likewise fairly quick. With practice, 10 image sequences can be analyzed for persistence length in an afternoon or so with a modern PC. The rate-limiting step in the analysis is the time to track individual fluorophores; newer tracking methods promise to increase this speed substantially27.
Because the method involves tracking single fluorophores attached to a given microtubule, the precision of tracking is only limited by photon collection, and can be on the scale of nanometers22,25 . By including data from many fluorophores, the precision is further improved. Potentially, even higher precision could be reached by attaching very bright fluorescent objects, such as quantum dots, to the microtubule.
While the precision of individual trajectories is quite good, the uncertainty in persistence lengths measured with this technique (on the scale of ± 50% for a single microtubule) is still significant. The key limitation in precision of the persistence length measurements is the statistics involved in averaging cosθs . As the contour length between pairs of points, s, increases, the number of independent measurements of θs decreases as L/s, where L is the length of the microtubule trajectory studied. Increasing the length of the microtubule trajectories would allow for improved precision in persistence length measurements.
A second limitation to this method is the requirement that biotinylated kinesin-1 be used for specific attachment of kinesin to microscope slides. Biotinylated kinesin must be purified (from E. Coli in our case), and cannot simply be purchased off the shelf. However, a modified gliding assay using unbiotinylated kinesin may be used28; kinesin heavy chain protein which might be suitable for this technique is commercially available from Cytoskeleton (KR01). Modifying the gliding assay to use this alternate kinesin would not affect microtubule flexural rigidity in any way, and would thus enable groups without access to recombinant kinesin protein to use this method.
Subscription Required. Please recommend JoVE to your librarian.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
We thank Melissa Klocke for assistance preparing Figure 1 and Anna Ratliff for demonstrating the protocol. This work was supported by the Research Corporation for Science Advancement.
Materials
| Name | Company | Catalog Number | Comments |
| Reagents | |||
| imidazole | Sigma-Aldrich | I2399 | |
| potassium chloride | Sigma-Aldrich | P9541 | |
| magnesium chloride | Sigma-Aldrich | M8266 | |
| EGTA | Sigma-Aldrich | E3889 | |
| BSA | Calbiochem | 126615 | |
| biotinylated BSA | Thermo Scientific | 29130 | |
| α-casein | Sigma-Aldrich | C6780 | |
| streptavidin | Thermo Scientific | 21125 | |
| dithiothreitol | Sigma-Aldrich | D0632 | |
| paclitaxel | LC Laboratories | P-9600 | |
| glucose oxidase | Sigma-Aldrich | G2133 | |
| catalase | Sigma-Aldrich | C100 | |
| glucose | Sigma-Aldrich | G8270 | |
| ATP | Sigma-Aldrich | A2383 | |
| 2-mercapt–thanol | Sigma-Aldrich | M3148 | Toxic. Buy small amount. |
| 24X60 mm No. 1 1/2 cover glass | VWR | 48393-252 | |
| 22X22 mm No. 1 cover glass | Gold Seal | 3306 | |
| High Vacuum Grease | Dow-Corning | NA | |
| Equipment | |||
| TIRF microscope | many | NA | The TIRF microscope used in this method was home-made. |
| IDL (software) | Exelis | NA | Could substitute MATLAB, ImageJ, or other image analysis software. |
References
- Hawkins, T., Mirigian, M., Selcuk Yasar, M., Ross, J. L. Mechanics of microtubules. Journal of biomechanics. 43, 23-30 (2010).
- Pampaloni, F., et al. Thermal fluctuations of grafted microtubules provide evidence of a length-dependent persistence length. Proceedings of the national academy of sciences U S A. 103, 10248-10253 (2006).
- Taute, K. M., Pampaloni, F., Frey, E., Florin, E. L. Microtubule dynamics depart from the wormlike chain model. Physical review letters. 100, 028102 (2008).
- Van den Heuvel, M. G., de Graaff, M. P., Dekker, C. Microtubule curvatures under perpendicular electric forces reveal a low persistence length. Proceedings of the national academy of sciences U.S.A. 105, 7941-7946 (2008).
- Gittes, F., Mickey, B., Nettleton, J., Howard, J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. Journal of cell biology. 120, 923-934 (1993).
- Brangwynne, C. P., et al. Bending dynamics of fluctuating biopolymers probed by automated high-resolution filament tracking. Biophysical journal. 93, 346-359 (2007).
- Cassimeris, L., Gard, D., Tran, P. T., Erickson, H. P. XMAP215 is a long thin molecule that does not increase microtubule stiffness. Journal of cell science. 114, 3025-3033 (2001).
- Valdman, D., Atzberger, P. J., Yu, D., Kuei, S., Valentine, M. T. Spectral analysis methods for the robust measurement of the flexural rigidity of biopolymers. Biophysical. 102, 1144-1153 (2012).
- van Mameren, J., Vermeulen, K. C., Gittes, F., Schmidt, C. F. Leveraging single protein polymers to measure flexural rigidity. Journal of physical chemistry b. 113, 3837-3844 (2009).
- Hua, W., Chung, J., Gelles, J. Distinguishing inchworm and hand-over-hand processive kinesin movement by neck rotation measurements. Science. 295, 844-848 (2002).
- Duke, T., Holy, T. E., Leibler, S. "Gliding assays" for motor proteins: A theoretical analysis. Physical review letters. 74, 330-333 (1995).
- Mehrbod, M., Mofrad, M. R. On the significance of microtubule flexural behavior in cytoskeletal mechanics. PLoS one. 6, e25627 (2011).
- Szarama, K. B., Gavara, N., Petralia, R. S., Kelley, M. W., Chadwick, R. S. Cytoskeletal changes in actin and microtubules underlie the developing surface mechanical properties of sensory and supporting cells in the mouse cochlea. Development. 139, 2187-2197 (2012).
- Gal, N., Weihs, D. Intracellular Mechanics and Activity of Breast Cancer Cells Correlate with Metastatic Potential. Cell biochemistry and biophysics. , (2012).
- Volokh, K. Y. On tensegrity in cell mechanics. Mol. Cell Biomech. 8, 195-214 (2011).
- Kasas, S., Dietler, G. Techniques for measuring microtubule stiffness. Current nanoscience. 3, 85-96 (2007).
- Kikumoto, M., Kurachi, M., Tosa, V., Tashiro, H. Flexural rigidity of individual microtubules measured by a buckling force with optical traps. Biophysical journal. 90, 1687-1696 (2006).
- Venier, P., Maggs, A. C., Carlier, M. F., Pantaloni, D. Analysis of microtubule rigidity using hydrodynamic flow and thermal fluctuations. Journal of biological chemistry. 269, 13353-13360 (1994).
- van den Heuvel, M. G., Bolhuis, S., Dekker, C. Persistence length measurements from stochastic single-microtubule trajectories. Nano letters. 7, 3138-3144 (2007).
- Phillips, R., Kondev, J., Theriot, J. Physical biology of the cell. , Garland Science. (2008).
- Berliner, E., Young, E. C., Anderson, K., Mahtani, H. K., Gelles, J. Failure of a single-headed kinesin to track parallel to microtubule protofilaments. Nature. 373, 718-721 (1995).
- Anderson, E. K., Martin, D. S. A fluorescent GTP analog as a specific, high-precision label of microtubules. Biotechniques. 51, 43-48 (2011).
- Hyman, A. Preparation of modified tubulins. Methods in enzymology. 196, 478-485 (1991).
- Lee, I. Curve reconstruction from unorganized points. Computer aded geometric design. 17, 161-177 (2000).
- Yildiz, A. Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science. 300, 2061-2065 (2003).
- Friedman, L. J., Chung, J., Gelles, J. Viewing dynamic assembly of molecular complexes by multi-wavelength single-molecule fluorescence. Biophysical. 91, 1023-1031 (2006).
- Smith, C. S., Joseph, N., Rieger, B., Lidke, K. A. Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nature Methods. 7, 373-375 (2010).
- Nitzsche, B., Ruhnow, F., Diez, S. Quantum-dot-assisted characterization of microtubule rotations during cargo transport. Nature. 3, 552-556 (2008).



