FIM Imaging and FIMtrack: Two New Tools Allowing High-throughput and Cost Effective Locomotion Analysis
Summary
FIM is a novel, cost effective imaging system designed to track small moving objects such as C. elegans, planaria or Drosophila larvae. The accompanying FIMTrack program is designed to deliver fast and efficient data analysis. Together, these tools allow high-throughput analysis of behavioral traits.
Abstract
The analysis of neuronal network function requires a reliable measurement of behavioral traits. Since the behavior of freely moving animals is variable to a certain degree, many animals have to be analyzed, to obtain statistically significant data. This in turn requires a computer assisted automated quantification of locomotion patterns. To obtain high contrast images of almost translucent and small moving objects, a novel imaging technique based on frustrated total internal reflection called FIM was developed. In this setup, animals are only illuminated with infrared light at the very specific position of contact with the underlying crawling surface. This methodology results in very high contrast images. Subsequently, these high contrast images are processed using established contour tracking algorithms. Based on this, we developed the FIMTrack software, which serves to extract a number of features needed to quantitatively describe a large variety of locomotion characteristics. During the development of this software package, we focused our efforts on an open source architecture allowing the easy addition of further modules. The program operates platform independent and is accompanied by an intuitive GUI guiding the user through data analysis. All locomotion parameter values are given in form of csv files allowing further data analyses. In addition, a Results Viewer integrated into the tracking software provides the opportunity to interactively review and adjust the output, as might be needed during stimulus integration. The power of FIM and FIMTrack is demonstrated by studying the locomotion of Drosophila larvae.
Introduction
Most animals have the ability to move in a highly sophisticated and controlled manner. To decipher the genetic basis underlying locomotion control it is mandatory to quantitatively assess different behavioral patterns. In this respect, Drosophila can serve as an ideal model. Tracking of freely flying Drosophila is tantalizing 1-4 but crawling of Drosophila larvae occurs in two dimensions at relatively low speed and can thus be monitored easily. Camera-based setups combined with appropriate illumination are used to acquire images 5. Both incident or transmitted light is employed in behavioral experiments 6,7. However, due to the semi-translucent body of the larvae and possible light reflections of the crawling surface faithful recording of larval movements can be challenging. To overcome such problems, some complex methods have been devised. Recently, dark field illumination was introduced to enhance the foreground/background contrast 8. As an alternative to camera-based recording, lens-less optical imaging and image-sensor-less on-chip acquisition techniques have been introduced 9-11.
Several tracking programs have been introduced recently, including commercially available software 12 and custom solutions. Examples for high-throughput tracking programs are the Multi Worm Tracker (MWT) 13 and Multianimal Gait And Track (MAGAT) 8. Both have in common, that multiple animals can be tracked in a single open-field arena so that colliding animals lead to multiple new animal identities. To overcome this limitation, a multi-well setup was introduced separating 12 animals into individual wells 14. Precise quantification of locomotion of single individuals can be achieved by using a movable tracking stage in combination with a microscope 15. However, all these approaches are either cost inefficient, lack sufficient resolution or too time consuming for high throughput phenotyping.
To overcome the above mentioned limitations, we have developed FIM (FTIR-based Imaging Method) based on Frustrated Total Internal Reflection (FTIR) 16 (Figure 1). This new imaging approach provides an unprecedented high contrast and even allows multi-color recording of crawling animals 16. The underlying principle of this handy and effective method is easy. An acrylic glass plate is flooded with light (e.g., 875 nm infrared). Due to different refractive indices of acrylic glass and air, the light is totally reflected at the glass/air boundary. No heating of the acrylic glass is noted 16. Only if objects with a higher refractive index touch the light-flooded table, can light enter these objects. If the animals touch the surface, light is reflected and can be captured from below (Figure 1). In consequence, only the contact area of the animals appears as a bright spot, which allows detailed imaging with an overall black background. Thus, FIM-imaging allows to record perfect movies for computer vision algorithms. The simple and robust use of FIM now brings detailed high throughput analysis of complex animal behavior into reach and can be used for studying information processing: e.g., olfaction 8, 16; vision 17 or thermosensation 18.
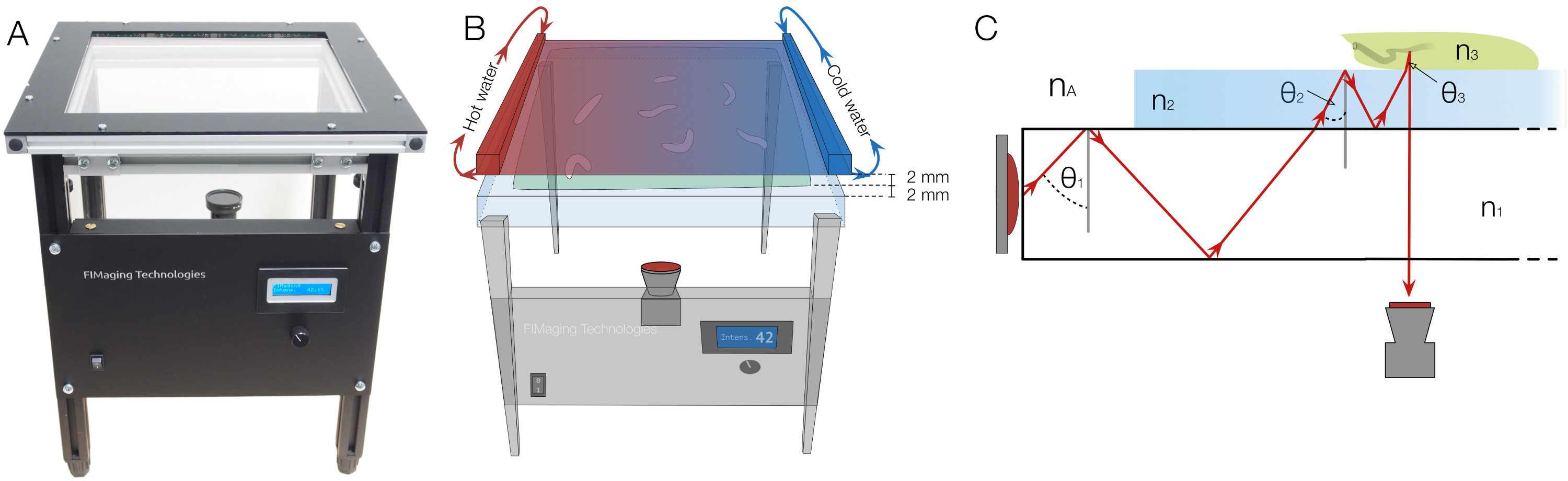
Figure 1. FIM setup with heat-stimulus integration and underlying physical principles. (A) The FIM setup. Illumination intensity can be regulated at front panel. (B) To deliver a heat stimulus, a black painted aluminum plate, perfused with hot and cold water at both sides, is placed 2 mm above the agar surface which itself is 2 mm thick. The gradient is established on the heat radiator plate and the agar by the temperature differences. (C) The physical principle of frustrated total internal reflection: An acrylic glass plate is illuminated by infrared light. θ1, θ2, and θ3 indicate the light reflection angles. nA, n1, n2 and n3 denote the refractive indices of air, acrylic glass, agar and the larva respectively and fulfill the inequality nA <n1 <n2 <n3. Due to refraction, the reflection angle changes during transition. If the angle is below the critical angle, light is not reflected anymore, can pass through the layers and can be captured from below. Please click here to view a larger version of this figure.
The spectrum of processes that can be analyzed by FIM is broad. Without any further adjustments, FIM imaging can be used to monitor all larval stages of Drosophila (Figure 5B) or can be used to follow the foot-prints of adult Drosophila 19. Likewise, the trajectories of C. elegans or the movement of planarian flatworms can be easily recorded (Figure 5C). Even the analysis of fungal hypha or root hair growth appears feasible 19. In our current FIM setup, 4 x 16 infrared light emitting diodes (IR- LEDs) are integrated into a 32 x 32 cm2 acrylic glass plate, called the tracking table (Figure 1). The intensity of the IR-LEDs is adjusted depending on the weight of the objects on the tracking table, which can be easily done by a micro controller connected to the circuit via pulse-width modulation (PWM). FIM yields very high contrast images over a broad range of illumination intensities. Importantly, it generates excellent results at already low overall infrared irridation.
A camera with an infrared filter is placed below the tracking table, which allows integration of additional stimuli into the setup. Heat stimuli can be easily applied by a heat radiator plate and light stimuli are applied by an LCD projector. Also odorants can be contained in gradients by simple lids 8. For heat gradient experiments, the heat radiator plate is perfused with hot and cold water on both sides respectively and placed 2 mm above the larvae (Figure 1B).
The generation of high contrast, high quality movies opens the possibility for sophisticated computer based image analysis, thus we implemented the FIMTrack software to extract a large set of features from images (Figure 2). First six primary features were defined from the contour of the animal (Figure 3A). These features provide the baseline for further calculation of six secondary features which describe the animals shape and its position at certain stimuli at a given time point (Figure 3B). Currently, nine tertiary features are calculated that are integrating temporal aspects and thus characterize the locomotion of the animal together with the primary and secondary features (Figure 3C).
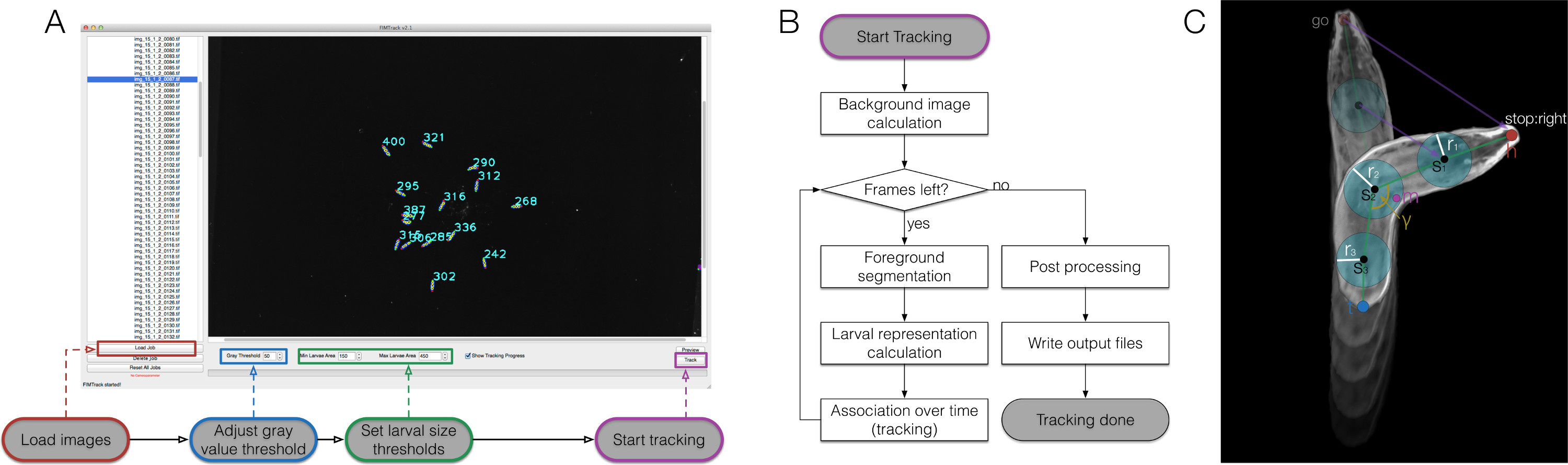
Figure 2. FIMTrack overview, algorithmic workflow and larval representation. (A) How to use FIMTrack. The images are loaded. Gray value threshold and larval size thresholds defining single larvae must be set. The larval area must be in [min-size, max-size]. Tracking is started by the highlighted button. (B) Tracking workflow. After the start button is clicked, the background image is calculated (minimal intensities over time). As long as there are frames left, the larvae are segmented based on the gray threshold and the min- and max-size threshold. For all segmentations the larval representations are calculated (compare to (C)). Each new model is associated to a given trajectory if a valid track is available. If the last frame is reached, finalizing post processing is done followed by the output generation. (C) Larval representation. The animal consists of a head and a tail point (h and t). Between these points an arbitrary odd number of spine points si can be set with a radius ri. In addition, the center of mass m and the main body bending angle γ are calculated. Several motion related parameters are sketched by purple lines. Please click here to view a larger version of this figure.

Figure 3. Features calculated by FIMTrack. (A) Primary features based on the contour of the animals. (B) Secondary features, based on primary features. (C) Tertiary features, based on primary features in consecutive frames and additional inputs Please click here to view a larger version of this figure.
Protocol
NOTE: Here, the use of FIM is presented for handy high throughput analysis of larval locomotion for screening in free-moving conditions and under influence of a heat stimulus. Other applications such as analysis of olfactory stimuli dependent locomotion or high resolution imaging of rolling or other behaviors might need subtle changes to the protocol, which are provided upon request.
1. Set-up of Experiments
- Use about 100 larvae per genotype in total (1 hr time required) to obtain statistically meaningful values. NOTE: In case several genotypes need to be compared and recorded on different days, record the same number of larvae per genotype per day.
- For most applications, record at 10 Hz for parameter analysis. For high resolution imaging applications, vary imaging speed if needed.
- For tracking of third instar larvae, conduct experiments about 120 hr after egg laying. Make sure that cultures yield enough larvae in the experimental period (see 4.3).
- For non-stimulus applications, prepare a crawling surface containing translucent agar (see section 2). To contain larvae in the stimulus range for heat gradient application, surround the field of view with an aversive salt agar barrier (see section 3).
NOTE: Other applications may require different surfaces. - Make sure to use a room with constant environmental conditions (temperature, light, air flow, air humidity etc.).
2. Crawling Surface Preparation (Translucent Agar)
NOTE: In the FIM setup an agar surface is added to provide a moist crawling surface. In addition, it also improves illumination properties.
- Boil 0.8% food grade agar in deionized ultrapure water. For screening application prepare 400 ml.
- Pour agar at 50 °C (hand-hot) on a separate 33 cm x 33 cm acrylic-glass plate. The surface tension of the agar and thus the temperature will define the thickness of the agar slab. At 50 °C, 2 mm thick agar slabs are obtained. Do not agitate after boiling and pour constantly to avoid bubbles. Prepare fresh agar slabs for every imaging period of about 4 hr.
- Fill standard 6 cm petri dishes with the remaining 0.8% agar solution to sort, clean and habituate larvae prior to the experiment (1 dish per genotype).
- In case an aversive agar barrier is needed for the application, proceed to section 3.
- Trim off approximately 2 cm of the perimeter of the agar slab to obtain a plane square surface for recording. Remove the excess agar.
NOTE: The size is dependent on the application. - Transfer the agar slab to the FIM setup directly after cooling by gently pushing the gliding agar over the edge of the acrylic glass plate.
3. Optional: Adding an Aversive Agar Barrier to the Crawling Surface (Salt Agar)
- Boil 2.5% food grade agar with 3 M NaCl in deionized ultrapure water. For screening in a heat gradient prepare 200 ml.
NOTE: The volume depends on the application. - Cut a 2 cm wide notch in the previously poured crawling surface surrounding the field of view (22 x 22 cm).
NOTE: Different barrier forms and fields of view may be required depending on the application. - Fill the notch with salt agar 0.1-0.3 cm higher than the tracking surface.
4. Fly Handling
- Rear flies in 25 °C incubator with 12 hr light/dark cycle adjusted to the experiment’s time on standard fly food at 65% air humidity.
- For crosses, anesthetize 30 virgin female flies and 8 males with CO2 and cross them in 130 ml culture vials with 35 ml food.
NOTE: One 130 ml culture vial with 35 ml food will yield about 20-30 late third instar larvae within the imaging span of 4 hr. Imaging and tracking works for all larval instar stages. - Drop a little water in culture vials to drive late third instar larval movement and collect the largest larvae from vial walls using a small paintbrush.
- 2-5 min before recording, transfer larvae for one video to a Petri-dish containing 0.8% food grade agar in deionized ultra pure water to habituate and clean them.
5. Adjusting the FIM Imaging Setup for Recordings (Non Stimulus Conditions)
- Adjust camera lens focus and aperture if needed. Set the exposure time for the respective camera.
NOTE: These settings do not need to be changed during an experiment. - Adjust illumination intensity to obtain good contrast by imaging one larva.
- Keep environmental conditions in the room constant during recordings. Darken the room to not disturb larvae with directional light.
6. Optional: Adjusting the FIM Imaging Setup for Recordings (Gradual Temperature Stimulus Conditions)
NOTE: The heat gradient device is a 42 x 42 x 0.2 cm3 aluminum plate with a matt black paint and isolating material on the top site. The plate is perfused with water from two different circuits connected to calorifiers/coolers and pumps at both ends (see Figure 1B). The temperatures can be regulated from -5 to +50 °C.
- Turn on the heat gradient device 1 hr prior to the experiments and place it above the setup to allow establishing the desired temperature profile and components to equilibrate.
- Transfer the crawling surface agar with a salt barrier to the setup.
- Place the radiator plate over the agar and adjust the spacing between the plate and the crawling surface to 2 mm (~1 mm above larvae).
- Establish a linear gradient of at least 0.8°C/cm from 34 °C (approx. 2 cm from the barrier in the field of view) to 18 °C (approx. 2 cm from the barrier at the opposite site in the field of view) by adjusting the temperature of the water circuits to 1 °C and 45 °C.
NOTE: Different gradient properties of the metal plate require different temperatures which need to be ascertained empirically. These settings do not need to be changed during the experiments. - Adjust settings as described in section 5.
- Allow the crawling surface to equilibrate in the gradient before experiments for 20 min. Test the temperature gradient with a pyrometer.
- Proceed with section 8.
7. FIM Imaging (Non Stimulus Conditions)
- Use a small wet paintbrush to gently gather 15 larvae from the habituation petri dishes and transfer them to the center of the tracking surface. Don’t transfer food remains or too much water (see 7.8).
- Record 1 – 50 larvae on a 22 cm x 22 cm tracking area. Use 15 animals per video for most screening applications (for statistical reasons use at least 100 individuals per genotype).
- Gently separate larvae and wait for about 10-20 sec until all larvae started to move straight before recording.
- Record larval locomotion for 2 min at 10 frames per sec for non stimulus conditions. Use uncompressed tif images as the output format.
- During recording, collect larvae for the next video from vials to habituate them. Critical: Do not disturb recorded larvae with light.
- After recording, remove larvae with a large paintbrush and discard them in accordance with local safety and legislation rules.
- After removing larvae, use the paintbrush to clean and moisturize the crawling surface with ultra pure deionized water.
- Critical: Keep the surface moist at all times but avoid excessive moisture, which can be seen as halos or drops surrounding larvae in recordings and disturb tracking.
8. Optional: FIM Imaging (Gradual Temperature Stimulus Conditions)
- After the temperature gradient is established (see section 6), prepare the recording software for instant recording within 20 sec after placing the larvae on the crawling surface (see 8.2) e.g., set the number of frames per video and define the saving path.
- Lift the radiator plate slightly and place the larvae at 33 °C 2 cm from the salt barrier. Refer to 6. and 7. for further general instructions. Lower the radiator plate again and start recording for 3-4 min directly after larvae started to move straight.
- After recording, remove larvae directly, clean and moisturize the surface. Do not touch the salt agar to avoid spreading of NaCl.
- Critical: Keep the surface moist at all times but avoid excessive moisture, which can be seen as halos or drops surrounding larvae in recordings and disturb tracking.
- Allow the agar surface to equilibrate again after moisturizing for 1-2 min, while saving images and gathering new animals before preparing for next video. Control the temperature gradient using a pyrometer every 5 videos and adjust temperature device if needed (water temperature, height, and x-y orientation in respect to the tracking surface).
9. Tracking of Larval Locomotion
NOTE: For more details refer to the attached manual for FIMTrack (supplement). For a program flow chart see Figure 2.
- Adjust tracking parameters for the respective experiments using the preview option.
- Adjust pixel per cm according to camera and field of view.
- Adjust frames per sec based on the camera settings.
- Adjust brightness thresholds so that all animals are detected correctly (feedback is given in the preview).
- Adjust larval area size thresholds. Note the feedback option in the preview: single animals are highlighted in yellow, colliding larvae are highlighted in red and the area of each animal is given in blue.
- Start tracking by using the button in the bottom right.
NOTE: After successful tracking, an image featuring larval tracks and a csv file containing the calculated locomotion and posture features is stored in the image directory. - Use the FIM Results viewer module (Edit >Results Viewer…) to review and manually adjust the tracking results. If required, define stimulus regions to evaluate data in respect to the stimulus regime e.g., crawling orientation in respect to the heat gradient or distance to an odorant source. For more detailed description please use the manual (supplement).
10. Evaluation of the Data
- Import the csv file into Excel, Matlab or any other program for further statistical analysis.
Representative Results
For imaging several different cameras with different resolution properties were tested (Figure 4). All cameras where equipped with an appropriate IR filter. According to the low resolution of the lowest priced camera in this test, the field of view is limited to 10 cm x 10 cm. Best results were obtained using a 4 megapixel (MP) camera. This leads to a resolution of 100 pixels per third instar larval length and allowed to easily recognize internal structures. Additionally, the peristalsis of the animal can be easily extracted (Figure 4A). However, one can still obtain high contrast movies using less expensive cameras, which can also be analyzed by FIMTrack. Using a 1.4 MP camera with a depth of 8 bit and a resolution of 1,392 x 1,040 pixels is about half the price and allows a resolution of 45 pixels per third instar larval length in the field of view. The head but no other internal structures can be recognized (Figure 4B). Tracking and detection of peristalsis is possible but accuracy is reduced (Figure 4B).
With an even cheaper 0.8 MP camera with a spatial resolution comparable to the 1.4 MP camera, the larval head cannot be recognized faithfully anymore (Figure 4C). Tracking and peristalsis analysis is possible but including more jitter based on the increased noise. Surprisingly, even a low resolution USB webcam provides sufficient quality movies to calculate larval trajectories (0.3 MP camera, below 20€, Figure 4D). The peristalsis can be calculated from the area but measurements are very noisy.
In our setup we routinely use the 4 MP camera. For screening, this camera allows monitoring a 22 cm x 22 cm arena, which obviously provides the opportunity to analyze large numbers of animals simultaneously so that high throughput analysis is feasible. Using this setting, larval length is represented by 40 pixels which still allows recording and analysis of peristalsis. An exemplary image of 15 larval trajectories in a heat gradient is given in Figure 5A. In addition, the use of a macro lens allows to image larvae with very high resolution where many internal organs become visible and head recognition is further improved (Figure 5B). Moreover these can be used for further more detailed analysis of behaviors of a wide range20. The same setup can be easily used to image crawling C. elegans worms (Figure 5C).

Figure 4. FIM imaging and tracking results for different cameras. (A) Left: FIM imaging of three larvae crawling on a 10 cm x 10 cm tracking stage captured using a 4 MP camera with 10 fps. Mid: Clipping and center of mass trajectory of a single larva. The area of the animal is indicated. Right: Area of the larva plotted over 100 frames. The red arrow indicates the time point of the clipped image. (B) Equivalent to (A) but captured using a 1.4 MP camera. (C) Equivalent to (A) but captured using a 0.8 MP camera. (D) Equivalent to (A) but captured using a 0.3 MP camera. Please click here to view a larger version of this figure.

Figure 5. Heat stimulus, and high resolution applications. (A) Heat stimulus application (compare to Figure 1). Trajectories calculated via FIMTrack. (B) High resolution application image of a third, second and first instar larva using a macro lens. The third instar larval length is represented by 400 pixels in a field of view of 2.5 x 2.5 cm. (C) A C. elegans worm captured using FIM imaging. Scale bars are indicated. Please click here to view a larger version of this figure.
Discussion
In behavioral neuroscience it is mandatory to quantitatively decipher complex behavioral traits. Thus, large numbers of individuals must be observed at high resolution and automated procedures are needed for statistical analysis. Here, FIM imaging is described, a novel, simple and robust imaging setup, which provides the means to monitor locomotion of a wide range of animals. The efficacy of the FIM imaging setup was tested using Drosophila larvae, planarian flatworms and C. elegans worms. The FIM technology provides intrinsically high contrast to detect even internal structures of the animals, such as the brain, trachea, the gut or the proventriculus. Importantly, these internal structures are robustly identified so that they can serve to automatically identify the orientation of the animal19.
The quality of the movies can be influenced by excessive amount of water on the crawling surface. Thus, it is critical to control the moisture of the agar. Too old agar or too much water on the surface can cause artifacts. Likewise, ensure that no air bubbles are included in the crawling surface. In general, a well-prepared agar surface allows recording movies for 4 hr.
Due to the underlying physical principles FIM imaging generates almost noise free image recordings, resulting in a superb image quality. This in turn facilitates subsequent computer-based image analysis and enables high throughput. However, the methodology is restricted to analyzing animals that directly contact the agar surface. The tracking software is challenged by animals forming a donut shape. Although a binary indicator recognizes the donut shape, a wrong spine might be calculated.
Due to the modular construction of the tracking table dual and triple color imaging is in reach. Moreover, additional stimuli (light, odorants, electrical or mechanical stimuli) can easily be delivered from above. The FIMTrack program designed to match the power of FIM imaging can be easily adopted to track Drosophila larvae, C. elegans or planarians. Thus and due to its straightforward and cheap construction (see http://FIM.uni-muenster.de), FIM imaging is feasible for a wide range of biomedical applications and in particular allows urgently needed high throughput studies.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
We are thankful to S. Thomas who has initiated this project, J. Hermann and U. Burgbacher for help in the construction of the FIM setup. This work was funded by the DFG (SFB 629 B6).
Materials
| Name of the Material/Equipment | Source | Catalog Number | Comments |
| FIM setup | Custom | details for construction or purchase of setups is available upon request | |
| Acrylic glass plate | Custom | Additional for agar pouring | |
| Heat radiator plate | Custom | Aluminum plate (paintet in matt black) perfusable on opposing sites with adjustable mounting | |
| Water calorifier/cooling pumps and hoses | Custom | based on GE healthcare MultiTempIII (No.: 18-1102-78) and Dr Bruno Lange GmBH (Typ: LTG013) | |
| Standard Camera (4 MP) | Basler | acA2040-25gm | Camera defaultly used for the FIM setup |
| Test Camera (1.4 MP) | QImaging | 1394 firewire (01- QIC-F-M-12 MONO) | Camera used for comparison |
| Test Camera (0.8 MP) | Point Grey | Dragonfly 2 (DR2-13S2M/C-CS) | Camera used for comparison |
| Test Camera (0.3 MP) | Sony | PS Eye USB2.0 camera | Camera used for comparison |
| Computer | Custom | equipped with at least i5 Intel processor or better, 16 GB RAM and sufficient HDD storage space [>1TB] | |
| Standard Fly food | Custom | ||
| Standard Fly vials 135 ml | Sarstedt AG&Co, Nümbrecht, Germany | 78,895 | |
| Petri dishes 9cm | Sarstedt AG&Co, Nümbrecht, Germany | 821,473 | |
| Ultrapure deionized water | Merck Millipore, Darmstadt, Germany | Synergy | |
| NaCl | Carl Roth GmbH, Karlsruhe, Germany | 3957.2 | |
| Food grade agar | AppliChem GmbH, Darmstadt, Germany | A0917,5000 | |
| Paintbrush (small and large) | Milan | Aquarell 310 Size 0 and 2 | |
| Pyrometer | Trotec | BP20 |
Referenzen
- Maimon, G., Straw, A. D., Dickinson, M. H. A Simple Vision-Based Algorithm for Decision Making in Flying Drosophila. Current Biology. 18 (6), 464-470 (2008).
- Frye, M. A., Dickinson, M. H. Closing the loop between neurobiology and flight behavior in Drosophila. Current opinion in neurobiology. 14 (6), 729-736 (2004).
- Fry, S. N. The Aerodynamics of Free-Flight Maneuvers in Drosophila. Science. 300 (5618), 495-498 (2003).
- Risse, B., Berh, D., Tao, J., Jiang, X., Klette, R., Klämbt, C. Comparison of two 3D tracking paradigms for freely flying insects. EURASIP Journal on Image and Video Processing. 2013 (1), 57 (2013).
- Yilmaz, A., Javed, O., Shah, M. Object tracking: A Survey. ACM Computing Surveys. 38 (4), (2006).
- Pistori, H., et al. Mice and larvae tracking using a particle filter with an auto-adjustable observation model. Pattern Recognition Letters. 31 (4), 337-346 (2010).
- Ramot, D., Johnson, B. E., Berry, T. L., Carnell, L., Goodman, M. B. The Parallel Worm Tracker: a platform for measuring average speed and drug-induced paralysis in nematodes. PloS one. 3 (5), e2208 (2008).
- Gershow, M., et al. Controlling airborne cues to study small animal navigation. Nature Methods. 9 (3), 290-296 (2012).
- Cui, X., et al. Lensless high-resolution on-chip optofluidic microscopes for Caenorhabditis elegans and cell imaging. Proceedings of the National Academy of Sciences of the United States of America. 105 (31), 10670-10675 (2008).
- Heng, X., et al. Optofluidic Microscopy – a Method for Implementing a High Resolution Optical Microscope on a Chip. Lab on a chip. 6 (10), 1274-1276 (2006).
- Liu, P., Martin, R. J., Dong, L. Micro-electro-fluidic grids for nematodes: a lens-less, image-sensor-less approach for on-chip tracking of nematode locomotion. Lab on a chip. 13 (4), 650-661 (2013).
- Spink, A. J., Tegelenbosch, R. A., Buma, M. O., Noldus, L. P. The EthoVision video tracking system–a tool for behavioral phenotyping of transgenic mice. Physiology. 73 (5), 731-744 (2001).
- Swierczek, N. A., Giles, A. C., Rankin, C. H., Kerr, R. A. High-throughput behavioral analysis in C. elegans. Nature methods. 8 (7), 592-598 (2011).
- Yu, C. -. C. J., Raizen, D. M., Fang-Yen, C. Multi-well imaging of development and behavior in Caenorhabditis elegans. Journal of neuroscience methods. 223, 35-39 (2014).
- Wang, S. J., Wang, Z. -. W. Track-A-Worm, An Open-Source System for Quantitative Assessment of C. elegans Locomotory and Bending Behavior. PloS one. 8 (7), e69653 (2013).
- Gomez-Marin, A., Stephens, G. J., Louis, M. Active sampling and decision making in Drosophila chemotaxis. Nature communications. 2, 441 (2011).
- Kane, E. A., et al. Sensorimotor structure of Drosophila larva phototaxis. Proc. Natl. Acad. Sci. U.S.A. 110 (40), E3868-E3877 (2013).
- Luo, L., et al. Navigational decision making in Drosophila thermotaxis. Journal of Neuroscience. 30 (12), 4261-4272 (2010).
- Risse, B., Thomas, S., Otto, N., Löpmeier, T., Valkov, D., Jiang, X., Klämbt, C. FIM, a Novel FTIR-Based Imaging Method for High Throughput Locomotion Analysis. PLoS one. 8 (1), e53963 (2013).
- Risse, B., Otto, N., Jiang, X., Klämbt, C. Quantifying subtle locomotion phenotypes of Drosophila larvae using internal structures based on FIM images. Comput Biol Med. 14, (2014).

