Summary
The present report describes an in vitro enzymatic assay to measure phosphatidylethanolamine methyltransferase activity using Leishmania cell extracts. This assay is based on the transfer of a radioactive methyl group from S-[Methyl-3H]adenosyl-L-methionine onto endogenous phosphatidylethanolamine.
Abstract
Phosphatidylethanolamine methyltransferases are biosynthetic enzymes that catalyze the transfer of one or more methyl group(s) from S-adenosyl-L-methionine onto phosphatidylethanolamine, monomethyl-phosphatidylethanolamine, or dimethyl-phosphatidylethanolamine to give either monomethyl-phosphatidylethanolamine, dimethyl-phosphatidylethanolamine or phosphatidylcholine. These enzymes are ubiquitous in animal cells, fungi, and are also found in approximately 10% of bacteria. They fulfill various important functions in cell physiology beyond their direct role in lipid metabolism such as in insulin resistance, diabetes, atherosclerosis, cell growth, or virulence. The present manuscript reports on a simple cell-free enzymatic assay that measures the transfer of tritiated methyl group(s) from S-[Methyl-3H]adenosyl-L-methionine onto phosphatidylethanolamine using whole cell extracts as an enzyme source. The resulting methylated forms of phosphatidylethanolamine are hydrophobic and thus, can be separated from water soluble S-[Methyl-3H]adenosyl-L-methionine by organic extraction. This assay can potentially be applied to any other cell types and used to test inhibitors/drugs specific to a phosphatidylethanolamine methyltransferase of interest without the need to purify the enzyme.
Introduction
Phosphatidylethanolamine methyltransferase (PEMT) enzymes catalyze the covalent attachment of one or more methyl groups using S-adenosylmethionine (SAM) as the methyl group donor onto PE, monomethyl-PE or dimethyl-PE to give monomethyl-PE, dimethyl-PE and/or phosphatidylcholine (PC). These enzymes are almost ubiquitous in animal cells and fungi. They can also be found in some plants 1 and approximately 10% of bacteria, particularly those that interact with eukaryotes 2.
PEMTs are relevant to the biology of the cell not only by contributing to the production of PC, which is the main lipid class in animal cells, but also by fulfilling other important cellular functions. In mammals, PEMTs are mainly expressed in the liver where they are required for normal secretion of very low-density lipoprotein and they also contribute to diet-induced obesity 3, atherosclerosis 4, and insulin resistance 5. Additionally, mammalian PEMT are also expressed in adipocytes, although to lower levels, and participate in fat deposition 6,7. PEMT role in cancer development 8, apoptosis 9, and cell growth 10 have also been demonstrated. In bacteria, PEMT enzymes have been shown to be important for normal cell growth 2, virulence 2, and symbiosis with the host plant 11.
The goal and rationale of the present protocol is to measure PEMT activity from whole cell extracts without the need to purify the enzyme. Two distinct protocols have been developed to measure PEMT activity. The first and most common one measures the transfer of tritiated methyl group from radioactive SAM onto PE, which is the topic of this article. This protocol has been originally developed to measure PEMT activity from yeast 12 and mammalian cells (liver) 13 to gain an understanding of PC biosynthesis in these cells as well as to determine the specificity of these enzymes. Later, this technique has been applied to other cell types such as bacteria 2 (using a basic pH value for the assay though 15) and protozoan parasites 14. This technique can be used with whole cell extracts as well as purified enzyme, and can potentially be applied to any cell extract system. A non radioactive assay has also been designed that relies on the enzymatic quantification of S-adenosylhomocysteine, the transmethylation product of SAM 16. The latter assay may be more convenient as it does not involve radioactivity but it is only suitable for purified enzymes.
Subscription Required. Please recommend JoVE to your librarian.
Protocol
1. Cell Extract Preparation
- Grow the Leishmania cells in a sterile plastic bottle sealed with air tight cap at 26 ºC in a medium made of 1x M199 supplemented with 20 mM HEPES pH7.4, 100 U/ml penicillin, 100 µg/ml streptomycin, 5 µg/ml heme, 0.35 g/L NaCO2H, 0.1 mM adenine, and 2 µg/ml biopterin without shaking. Harvest the cells by centrifugation at 1,500 g for 5 min at 4 ºC when they reach a cell density of 1-2 x 107/ml.
- Discard the supernatant with a pipette and wash the cells by resuspending the cell pellet with a serological pipet in half of the culture's volume of cold phosphate buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH7.4). Dispose cell supernatants according to BL2 safety guidelines.
- Centrifuge cells again at 1,500 g for 5 min at 4 ºC. Discard the supernatant with a pipette. Proceed to the next step or snap freeze the cell pellet in liquid nitrogen for long term storage at -80 ºC (up to three months).
- Prepare 2x lysis buffer (0.5 M sucrose, 0.1 M TrisHCl, pH7.5, 2 mM EDTA, and 2x protease inhibitor cocktail) and keep it at 4 ºC on ice.
- Resuspend the cell pellet (fresh or frozen) in equal volume of 2x lysis buffer. Add 1x volume of glass beads. Vortex vigorously at 4 ºC for 10 min.
- Add 2 volumes of 1x lysis buffer and mix. Centrifuge cell extracts at 1,500 x g at 4 ºC for 10 min to pellet unbroken cells and nuclei.
- Transfer the supernatant with a pipet into a fresh cool centrifuge tube and keep cell extracts on ice until completion of the experiment.
2. Determine the Protein Concentration of the Cell Extract Using Protein Estimation Kit Such as Bicinchoninic Acid Assay
- Prepare the bicinchoninic acid (BCA) solution (1 ml/tube) by mixing the BCA and copper (II) sulfate in a ratio of 49:1 (v/v).
- Prepare the protein standards of 0, 10, 20, 30, 40, 50, and 60 µg/ml by diluting a 10 mg/ml bovine serum albumin (BSA) stock solution into 1 ml aliquots of the BCA solution.
- Add 2 µl of cells extracts in 1 ml of BCA solution in duplicates. Incubate standards and protein samples for 10 min in a pre-warmed 60 °C water bath.
- Transfer samples to ice for 3 min. Measure the absorbance of the standards and the protein samples with a spectrophotometer at a wavelength of 562 nm.
- Calculate the protein concentration of the cell extracts using the BSA standard as a reference as described in the manufacturer's protocol. Dilute cell extracts to a protein concentration of 10 mg/ml with 1x lysis buffer.
3. Enzymatic Assay in 200 μl per Tube
NOTE: Carry the following steps in a chemical hood.
- Test each sample in duplicate in a 15 ml conical tube. Prepare 20 μl 1 M TrisHCl pH 7.5 per tube and keep it on ice. Prepare 2 ml of chloroform/methanol (1:1 (v/v); stopping solution) at RT for each tube.
- Pipet 20 μl of 1 M TrisHCl pH 7.5 in each 15 ml conical tube on ice.
- Follow radiation safety guidelines from here on. Add the equivalent of 0.06 μM (0.2 μCi) S-[Methyl-3H]adenosyl-L-methionine and 50 μM cold SAM per tube for a total of 50.06 μM of SAM. Add x μl of cold water where x = 200-(20 (for buffer) + 20 (for cell extracts) + volume of cold and radioactive SAM) per tube.
- Transfer each conical tube to a pre-warmed 30 ºC water bath. Add to each tube 20 μl of cell extracts (equivalent of 200 μg of protein) to start the reaction. Incubate for the desired time (0 to 45 min).
- Stop the reaction by adding 2 ml of chloroform/methanol (1:1; v/v; stopping solution) to each tube. Transfer the conical tube to RT (20-25 ºC).
4. Lipid Extraction
NOTE: Carry the following steps in a chemical hood.
- Add 700 μl of water to each tube containing the enzymatic reaction sample. Vortex vigorously for 30 sec. Centrifuge at 1,500 x g for 5 min at RT to separate the organic from the water phase.
- Transfer the lower organic phase into a new 15 ml conical tube with a pipette. Add 1 ml of water to each "lower phase" containing tube and vortex vigorously for 30 sec. Centrifuge again at 1,500 x g for 5 min to separate the organic from the water phase.
- Transfer the lower organic phase into a scintillation tube with a pipette. Dry samples under a stream of N2. Dispose the water phases containing the non-incorporated radioactive SAM and the radioactive conical tubes as per radiation guidelines.
- Add 2 ml/tube of scintillation liquid. Measure the incorporated radioactivity with a scintillation counter according to the manufacturers' protocol and instrument's use.
- Calculate the enzymatic activity in nmol/mg protein using the following general equation:
cpm value x 103 x [total (radioactive and cold) concentration of SAM (mM)] x 5 specific activity of radioactive SAM (Ci/mmol) x [concentration of radioactive SAM (mM)]
NOTE: the above protocol can be altered for a SAM concentration dependent or a protein dependent PEMT assay. For SAM concentration dependent enzymatic assay, time is kept constant (15 min, which is in the linear range) and various amounts of cold SAM are added to the assay while for protein dependent PEMT assay, SAM concentration (we chose 0.05 mM) and time are kept constant (15 min). Also the pH of the buffer can be changed as needed if some PEMT enzymes have a different optimal pH value.
Subscription Required. Please recommend JoVE to your librarian.
Representative Results
Figure 1 shows a time dependant PEMT assay, which was carried out with Leishmania whole cell extract as an enzyme source using endogenous PE as a substrate. The amount of radioactivity in the organic phase was quantified by scintillation counting. The resulting numbers were utilized to calculate the amount of tritiated methyl groups transferred onto PE. The PEMT activity was linear for approximately 20 min. It then reached a plateau at around 30 min, after which it stayed constant for another 15 min. As expected, PEMT activity was not detected when no cell extracts were added to the assay (Figure 2). Further, this activity was abolished in the presence of 100 μM octadecyltrimethylammonium bromide, which is an inhibitor of L. major PE methyltransferases LmjPEM1 and LmjPEM2 14. PEMT activity was also protein concentration dependent, and this activity was linearly proportional to the amount of protein applied for the enzymatic assay (Figure 3). Lastly, a SAM concentration dependent PEMT assay was carried out, in which increasing concentrations of SAM were tested (Figure 4). PEMT activity reached a plateau at SAM concentration of approximately 0.5 mM. All together, these four assays demonstrate that PEMT activity is specific and can be measured from whole cell extracts without the need to purify the enzyme(s).
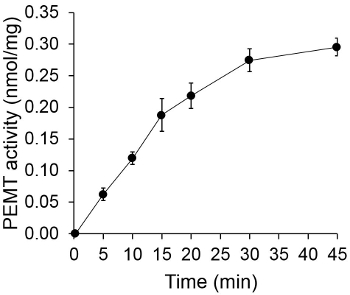
Figure 1. Time dependent PEMT assay. The enzymatic assay was performed twice in duplicate with 0.2 mg of whole Leishmania cell extracts as a function of time. PEMT activity is represented as nmol methyl groups transferred onto PE per mg of protein and per hr. For time "O", cell extracts were first mixed with 2 ml of stopping solution made of chloroform/methanol (1:1, by volume) before being added to the assay solution containing SAM. Standard deviations are shown.
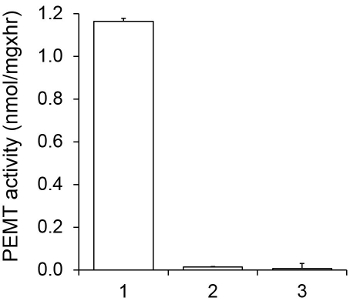
Figure 2. Specificity of PEMT activity. PEMT assay was carried out twice in duplicate for 15 min in the presence of 0.05 mM SAM. 1, 0.2 mg protein extract; 2, no cell extract; 3, 0.2 mg protein and 0.1 mM octadecyltrimethylammonium bromide. Standard deviations are shown.
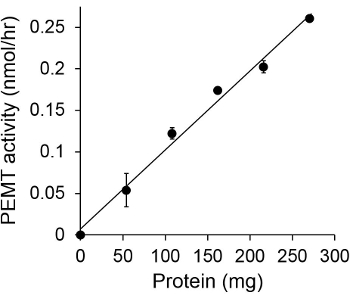
Figure 3. Protein dependent PEMT assay. The enzymatic assay was performed twice in duplicate in the absence (point 'O') or presence of various amounts of Leishmania proteins with 0.05 mM SAM for 15 min. Standard deviations are shown.
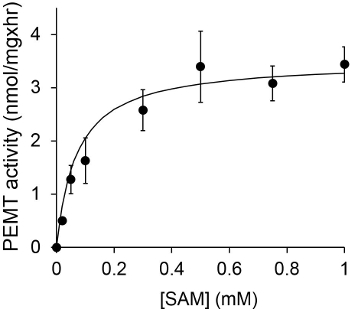
Figure 4. The PEMT assay was carried out twice in duplicate with 0.2 mg of whole Leishmania cell extracts in the presence of various concentrations of SAM for 15 min. Standard deviations are shown.
Subscription Required. Please recommend JoVE to your librarian.
Discussion
This simple, quick PEMT assay allows the quantification of methylated forms of PE that results from the transfer of radioactive methyl groups from SAM onto PE using whole cell extract as a protein source. It is fast, sensitive, reproducible, and also suitable for purified enzymes 17. Monomethyl- or dimethyl-PE can be added to the assay if the methyltransferase of interest is specific to these substrates rather than to PE 12,13,18,19. If purified PEMT enzyme is used, PE can be added to the assay. A limitation of this protocol is that the assay does not identify the reactions products (monomethyl-PE, dimethyl-PE, or PC). However, the identity of the reaction products (monomethyl-PE, dimethyl-PE, PC) can further be analyzed by one dimensional thin layer chromatography as described in 20,21. Further, some of the SAM degradation products such as S-adenosylhomocysteine and 5'-methylthioadenosine may inhibit PEMT activity by feedback inhibition. However, Leishmania possesses an S-adenosylhomocysteine hydrolase 22, which cleaves S-adenosylhomocysteine into adenine and S-ribosylhomocysteine, and a methylthioadenosine phosphorylase, which produces adenine and methylthioribose-1-phosphate 23. However, it is not known whether S-adenosylhomocysteine hydrolase and methylthioadenosine phosphorylase activities are high enough to efficiently metabolize S-adenosylhomocysteine and methylthioadenosine, respectively, so that no inhibition of PEMT activity occurs. In the instance S-adenosylhomocysteine hydrolase and/or 5'-methylthioadenosine metabolic enzyme are absent in the cell of interest, addition of the respective purified, recombinant enzymes can be added to the assay to relieve feedback inhibition by SAM degradation products 24,25,26.
There are four critical steps in this protocol: i) the protease inhibitor cocktail powder has to be added to the lysis buffer just before use (step 1.1); ii) the whole cell extracts are to be utilized within the following hours after preparation (after step 1.4); iii) during the lipid extraction steps (steps 4.2 and 4.4), caution needs to be applied to not transfer any of the interphase or water phase, which contains the excess tritiated SAM, and iv) to aliquot the radioactive S-[Methyl-3H]adenosyl-L-methionine and cold reagent upon receipt as repeated cycles of freeze and thaw degrade it into 5'-methylthioadenosine and homoserine lactone followed by hydrolysis to adenine and S-pentosylmethionine 26,27,28, which may account for lack of measurable PEMT activity. Bad whole cell extracts may also be responsible for no enzymatic activity. In this case, the quality of the cell extract can be assessed by measuring another known enzymatic activity. Last, lack of PEMT activity may rely on the PEMT enzyme being specific to monomethyl-PE or dimethyl-PE rather than PE. Addition of monomethyl-PE and/or dimethyl-PE substrates to the assay may restore PEMT activity.
An alternative, non-radioactive SAM dependent methyltransferase assay has been developed, that relies on the enzymatic quantification of homocysteine, the transmethylation products of SAM 29. This protocol has been successfully applied to measure PE specific methyltransferase activity 16. Unfortunately, this assay is only suitable for purified enzyme and not for whole cell extract, and does not reveal the identity of the reaction products either.
The present protocol can possibly be applied to any cell type. Additionally, this assay can be used to test potential drugs specific to a PEMT of interest in the context of testing new anti-microbial compounds to fight bacterial infections or novel therapeutics against diet-induced atherosclerosis, obesity, and insulin resistance.
Subscription Required. Please recommend JoVE to your librarian.
Disclosures
No conflict of interest declared
Acknowledgments
This work was supported by NIH grants ARRA RO3 AI078145 and 1SC3GM113743 to RZ.
Materials
| Name | Company | Catalog Number | Comments |
| S-[Methyl-3H]adenosyl-L-methionine (specific activity of 5-15 Ci/mMole) | Perkin Elmer | NET155050UC | Aliquot the reagent and freeze at -20 °C; follow radiation safety guidelines while using this reagent |
| Protease inhibitor cocktail | Roche Life Sciences | 11836170001 | dilute it fresh |
| Glass beads, acid washed, 425-600 mm | Sigma Aldrich | G8772 | |
| Bicinchoninic acid solution | Sigma Aldrich | B9643 | |
| Copper (II) sulfate | Sigma Aldrich | C2284 | |
| Scintillation counter MicroBeta2 with 1-detector | Perkin Elmer | 2450-0010 | |
| Spectrophotometer Biomate 3 | Thermo Scientific | 840208300 | |
| BSA stock solution (10 mg/ml) | New England Biolabs | B9001S | |
| Scintillation liquid | Research Product International Corp | 111198 | |
| S-(5'-Adenosyl)-L-methionine chloride (hydrochloride) | Cayman Chemicals | 13956 | dilute the reagent in 20 mM HCl and freeze aliquots at -80 °C |
References
- Keogh, M. R., Courtney, P. D., Kinney, A. J., Dewey, R. E. Functional characterization of phospholipid N-.methyltransferases from Arabidopsis and soybean. J Biol Chem. 284 (23), 15439-15447 (2009).
- Geiger, O., Lopez-Lara, I. M., Sohlenkamp, C. Phosphatidylcholine biosynthesis and function in bacteria. Biochim Biophys Acta. 1831 (3), 503-513 (2013).
- Gao, X., et al. Decreased lipogenesis in white adipose tissue contributes to the resistance to high fat diet-induced obesity in phosphatidylethanolamine N-.methyltransferase-deficient mice. Biochim Biophys Acta. 1851 (2), 152-162 (2015).
- Zhao, Y., et al. Lack of phosphatidylethanolamine N-.methyltransferase alters plasma VLDL phospholipids and attenuates atherosclerosis in mice. Arterioscler Thromb Vasc Biol. 29 (9), 1349-1355 (2009).
- Vance, D. E. Phospholipid methylation in mammals: from biochemistry to physiological function. Biochim Biophys Acta. 1838 (6), 1477-1487 (2014).
- Nishimaki-Mogami, T., Suzuki, K., Takahashi, A. The role of phosphatidylethanolamine methylation in the secretion of very low density lipoproteins by cultured rat hepatocytes: rapid inhibition of phosphatidylethanolamine methylation by bezafibrate increases the density of apolipoprotein B48-containing lipoproteins. Biochim Biophys Acta. 1304 (1), 21-31 (1996).
- Noga, A. A., Zhao, Y., Vance, D. E. An unexpected requirement for phosphatidylethanolamine N-.methyltransferase in the secretion of very low density lipoproteins. J Biol Chem. 277 (44), 42358-42365 (2002).
- Li, D., et al. Epigenetic repression of phosphatidylethanolamine N-.methyltransferase (PEMT) in BRCA1-mutated breast cancer. Oncotarget. 5 (5), 1315-1325 (2014).
- Cui, Z., Houweling, M., Vance, D. E. Suppression of rat hepatoma cell growth by expression of phosphatidylethanolamine N-.methyltransferase-2. J Biol Chem. 269 (40), 24531-24533 (1994).
- Cui, Z., Shen, Y. J., Vance, D. E. Inverse correlation between expression of phosphatidylethanolamine.N-.methyltransferase-2 and growth rate of perinatal rat livers. Biochim Biophys Acta. 1346 (1), 10-16 (1997).
- Minder, A. C., de Rudder, K. E., Narberhaus, F., Fischer, H. M., Hennecke, H., Geiger, O. Phosphatidylcholine levels in.Bradyrhizobium japonicum. membranes are critical for an efficient symbiosis with the soybean host plant. Mol Microbiol. 39 (5), 1186-1198 (2001).
- Kodaki, T., Yamashita, S. Yeast phosphatidylethanolamine methylation pathway. Cloning and characterization of two distinct methyltransferase genes. J Biol Chem. 262 (32), 15428-15435 (1987).
- Tanaka, Y., Amano, F., Maeda, M., Nishijima, M., Akamatsu, Y. Purification and properties of phosphatidyl-N-.monomethylethanolamine N-.methyltransferase, the enzyme catalyzing the second and the third steps in the phosphatidylethanolamine N-.methyltransferase system, from mouse liver microsomes. Jpn J Med Sci Biol. 43 (3), 59-73 (1990).
- Bibis, S. S., Dahlstrom, K., Zhu, T., Zufferey, R. Characterization of Leishmania major phosphatidylethanolamine methyltransferases LmjPEM1 and LmjPEM2 and their inhibition by choline analogs. Mol Biochem Parasitol. 196 (2), 90-99 (2014).
- deRudder, K. E., Thomas-Oates, J. E., Geiger, O. Rhizobium meliloti. mutants deficient in phospholipid N-.methyltransferase still contain phosphatidylcholine. J Bacteriol. 179 (22), 6921-6928 (1997).
- Aktas, M., Narberhaus, F. In vitro characterization of the enzyme properties of the phospholipid N-.methyltransferase PmtA from Agrobacterium tumefaciens. J Bacteriol. 191 (7), 2033-2041 (2009).
- Ridgway, N. D., Vance, D. E. Phosphatidylethanolamine N-.methyltransferase from rat liver. Methods Enzymol. 209, 366-374 (1992).
- Gaynor, P. M., Carman, G. M. Phosphatidylethanolamine methyltransferase and phospholipid methyltransferase activities from Saccharomyces cerevisiae. Enzymological and kinetic properties. Biochim Biophys Acta. 1045 (2), 156-163 (1990).
- Arondel, V., Benning, C., Somerville, C. R. Isolation and functional expression in Escherichia coli. of a gene encoding phosphatidylethanolamine methyltransferase (EC 2.1.1.17) from Rhodobacter sphaeroides. J Biol Chem. 268 (21), 16002-16008 (1993).
- Wessel, M., Klusener, S., Godeke, J., Fritz, C., Hacker, S., Narberhaus, F. Virulence of Agrobacterium tumefaciens. requires phosphatidylcholine in the bacterial membrane. Mol Microbiol. 62 (3), 906-915 (2006).
- Klusener, S., Aktas, M., Thormann, K. M., Wessel, M., Narberhaus, F. Expression and physiological relevance of Agrobacterium tumefaciens. phosphatidylcholine biosynthesis genes. J Bacteriol. 191 (1), 365-374 (2009).
- Henderson, D. M., et al. Cloning of the gene encoding Leishmania donovani.S.-adenosylhomocysteine hydrolase, a potential target for antiparasitic chemotherapy. Mol Biochem Parasitol. 53 (1-2), 169-183 (1992).
- Koszalka, G. W., Krenitsky, T. A. 5'-Methylthioadenosine (MTA) phosphorylase from promastigote of Leishmania donovani. Purine and Pyrimidine Metabolism in Man V, Adv Exp Med Biol. Nyhan, W. L., Thompson, L. F., Watts, R. W. E. 131, Springer US. 559-563 (1986).
- Biastoff, S., Teuber, M., Zhou, Z. S., Dräger, B. Colorimetric activity measurement of a recombinant putrescine N.-methyltransferase from Datura stramonium. Planta Med. 72 (12), 1136-1141 (2006).
- Hendricks, C. L., Ross, J. R., Pichersky, E., Noel, J. P., Zhou, Z. S. An enzyme-coupled colorimetric assay for S.-adenosylmethionine-dependent methyltransferases. Anal Biochem. 326 (1), 100-105 (2004).
- Cannon, L. M., Butler, F. N., Wan, W., Zhou, Z. S. A stereospecific colorimetric assay for (S.,S.)-adenosylmethionine quantification based on thiopurine methyltransferase-catalyzed thiol methylation. Anal Biochem. 308 (2), 358-363 (2002).
- Hoffman, J. L. Chromatographic analysis of the chiral and covalent instability of S.-adenosyl-L-methionine. Biochemistry. 25 (15), 4444-4449 (1986).
- Wu, S. E., Huskey, W. P., Borchardt, R. T., Schowen, R. L. Chiral instability at sulfur of S.-adenosylmethionine. Biochemistry. 22 (12), 2828-2832 (1983).
- Dorgan, K. M., et al. An enzyme-coupled continuous spectrophotometric assay for S.-adenosylmethionine-dependent methyltransferases. Anal Biochem. 350 (2), 249-255 (2006).



