Preparation of Hydroxy-PAAm Hydrogels for Decoupling the Effects of Mechanotransduction Cues
Summary
We present a new polyacrylamide hydrogel, called hydroxy-PAAm, that allows a direct binding of ECM proteins with minimal cost or expertise. The combination of hydroxy-PAAm hydrogels with microcontact printing facilitates independent control of many cues of the natural cell microenvironment for studying cellular mechanostransduction.
Abstract
It is now well established that many cellular functions are regulated by interactions of cells with physicochemical and mechanical cues of their extracellular matrix (ECM) environment. Eukaryotic cells constantly sense their local microenvironment through surface mechanosensors to transduce physical changes of ECM into biochemical signals, and integrate these signals to achieve specific changes in gene expression. Interestingly, physicochemical and mechanical parameters of the ECM can couple with each other to regulate cell fate. Therefore, a key to understanding mechanotransduction is to decouple the relative contribution of ECM cues on cellular functions.
Here we present a detailed experimental protocol to rapidly and easily generate biologically relevant hydrogels for the independent tuning of mechanotransduction cues in vitro. We chemically modified polyacrylamide hydrogels (PAAm) to surmount their intrinsically non-adhesive properties by incorporating hydroxyl-functionalized acrylamide monomers during the polymerization. We obtained a novel PAAm hydrogel, called hydroxy-PAAm, which permits immobilization of any desired nature of ECM proteins. The combination of hydroxy-PAAm hydrogels with microcontact printing allows to independently control the morphology of single-cells, the matrix stiffness, the nature and the density of ECM proteins. We provide a simple and rapid method that can be set up in every biology lab to study in vitro cell mechanotransduction processes. We validate this novel two-dimensional platform by conducting experiments on endothelial cells that demonstrate a mechanical coupling between ECM stiffness and the nucleus.
Introduction
Many aspects of the local cellular microenvironment (e.g., rigidity, pore size, nature of proteins, or cell-ligand density) provide a coordinate set of regulatory cues that control cellular processes such as motility, cell proliferation, differentiation, and gene expression. Modifications of the physicochemical properties of the extracellular environment can be perceived by cells and cause different physiological consequences, including deformation of cellular polarization, migration, and differentiation. It remains unclear, however, how cells translate ECM modifications into cellular biochemical signals. It is therefore of major importance to engineer controlled in vitro microenvironments that can reproduce the interactions between cells and their microenvironment for studying mechanotransduction pathways. To address this problem, we have recently introduced a novel method1, called hydroxy-PAAm hydrogels, to easily generate two-dimensional soft matrices that permit to independently control important mechanotransduction cues: matrix stiffness, cell geometry and confinement, nature of the protein and cell-ligand density.
ECM directs cellular processes via gradients in morphogens (chemotaxis), adhesive proteins (haptotaxis), and stiffness (durotaxis). Over the last few decades, advanced in vitro platforms have been developed to isolate these extracellular cues in order to tease out how cells are able to translate biochemical and biophysical features into physiological processes2-5. Electron-beam6, photolithography7, photochemical immobilization8, or plasma-assisted techniques9 have been developed to direct the growth of living cells on micropatterned substrates. Although these techniques have yielded important results, most of them do not allow discrimination between the individual influence of different cues on cell behavior and they require technical facilities that few laboratories can afford. Among these techniques, microcontact printing (µCP), has emerged as a robust and accessible method to create cell-adhesive micro-islands10. More recently, extensive efforts11-14 have been made to develop µCP on hydrogels with tunable rigidities in order to reproduce the wide range of rigidities observed in living tissues. Among these works, polyacrylamide (PAAm) has become popular15 and is already one of the most commonly used polymer-based matrices for cell biomechanics assays.
PAAm surfaces are commonly functionalized with the heterobifunctional cross-linker N-sulfosuccinimidyl-6-[4'-azido-2'-nitrophenylamido] (sulfo-SANPAH) and ECM proteins are linked to the surface by UV activation of the sulfo-SANPAH nitrophenyl azide groups16. Another technique consists in coupling hydrazine to proteins that have been severely oxidized with periodate17. Hynd and coworkers introduced a technique for patterning biomimetic hydrogel surfaces with protein and peptides that requires photopolymerization in presence of an acroyl-streptavidin monomer18. More recently, Tseng et al. have reported a new micropatterning method19 based on deep UV exposure of PAAm through an optical quartz mask that requires to incubate activated PA gels with 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) water solutions prior to add the protein. Despite the ability of these techniques to create homogeneous and reproducible proteins micropatterns, most of them suffer major limitations: long synthesis processes (e.g., dialysis, lyophilization, etc), expensive chemical compounds (e.g., hyaluronic acid, sulfo-SANPAH) or deep UV irradiation. In addition, these techniques do not allow independent modulation of substrate stiffness, micropattern geometry, ECM protein nature, and cell-ligand density.
Taking these limitations into account, we have developed a novel and simple acrylamide-based approach that allows immobilization of a variety of proteins and biomolecules on soft hydrogels and permits independent tuning of mechanotransduction cues in order to decipher their role on cellular functions. Instead of treating PAAm hydrogels with harsh chemical compounds, we introduce a commercial acrylamide monomer with hydroxyl groups during PAAm polymerization. This simple operation overcomes the intrinsic anti-adhesive property of PAAm hydrogels without any other technical requirements.
The presence of hydroxyl groups leads to a high affinity of hydroxy-PAAm hydrogels for proteins and biomolecules that form hydrogen-bonding interactions. In combination with µCP, hydroxy-PAAm hydrogels enable a rapid generation of two-dimensional culture platform with an independent control on matrix rigidity, type of ECM proteins, cell-ligand density and confined adhesiveness, which are envisioned to be a powerful platform for studying mechanotransduction.
The purpose of this protocol is to provide the necessary information for easily making hydroxy-PAAm hydrogels without any expertise in material sciences. The ultimate goal is to provide a means for researchers to ask physiologically relevant questions at the cellular and tissue levels that may lead to a better understanding of mechanotransduction pathways involved in pathophysiological mechanisms.
Protocol
1. Activating the Surface of Glass Coverslips
- Place glass circular coverslips (25 mm diameter) in a Petri dish and smear 0.1 M NaOH solution on it for 5 min (chemical fume hood recommended).
- Remove the NaOH solution and fully immerse coverslips with sterile ddH2O for 20 min while gently rocking on a rocking plate in a sterile culture hood.
- Drain sterile ddH2O and repeat the step 1.2.
- Remove the coverslips with sterile tweezers and place them in a new Petri dish with the activated face up.
- Dry coverslips under a steady flow of high-purity nitrogen gas.
- In a sterile culture hood, smear a thin layer of 3-(trimethoxysilyl)propyl acrylate (92%) on the activated side of the coverslip for 1 hr.
- Extensively wash glass coverslips with 3 washes of sterile ddH2O and immerse them in sterile ddH2O in a new Petri dish.
- Tap the Petri dish with Parafilm and place it on a rocker plate under gentle agitation for 10 min.
- Remove the coverslips from ddH2O with sterile tweezers with fine tips and place them in a new Petri dish with the activated face up.
- Store at RT in a dry place with aluminum foil to avoid dust from sticking to the coverslips.
2. Preparation of Hydroxy-PAAm Hydrogels
- Prepare a weight of 65 mg of N-hydroxyethyl acrylamide (HEA) in a 1.5 ml Eppendorf tube. It is important to prepare a fresh HEA solution.
- Add 1 ml of 50 mM HEPES buffer to HEA and mix using a vortexer until the complete HEA dissolution.
- Add 400 µl of 40% w/w in HEPES acrylamide solution and the required volume of 2% w/w in HEPES bis-acrylamide solution (see Table 1) to reach the desired hydrogel stiffness. Adjust with 50 mM HEPES to a final volume of 5 ml.
- Mix the solution using a vortexer and degas it in a vacuum chamber for 20 min in order to reduce oxygen concentration within the solution, which prevents hydroxy-PAAm polymerization.
- Under a sterile hood, filter the degased solution with a 0.2 µm pore size filter in order to sterilize it.
- Activate circular glass coverslips (22 mm diameter) in a UV/ozone cleaner during 7 min.
- Prepare 100 µl of 10% ammonium persulfate (APS) solution, that is 10 mg APS in 100 µl ddH2O. It is important to prepare a fresh APS solution.
- Add 2.5 µl of tetramethylenediamine (TEMED) and 25 µl of APS solution to the sterilized hydroxy-PAAm solution (step 2.5) to initiate the polymerization. Mix the solution by 3 successive pipettings without introduction of bubbles, under sterile conditions.
- Under a sterile hood, place a 25 µl drop of the hydroxy-PAAm solution on a 25 mm coverslip (available from step 1.9) and immediately place a 22 mm glass coverslip (prepared on step 2.5) on top of the droplet to squeeze the hydroxy-PAAm solution. Center the 22 mm glass coverslip with sterile tweezers and smooth out any bubbles.
- Allow hydroxy-PAAm hydrogels to polymerize at RT for 15 min. Invert manually the remaining hydroxy-PAAm solution in the Eppendorf tube to follow the completion of the polymerization process.
- Fully immerse coverslips with sterile ddH2O and carefully separate the 22 mm glass coverslips by introducing the edge of a razor blade between the 22 mm glass coverslips and the hydroxy-PAAm hydrogel layer.
- Wash hydroxy-PAAm hydrogels with sterile PBS (3 exchanges of PBS) and let the gels fully immersed in sterile PBS to maintain hydration.
- Store hydroxy-PAAm hydrogels in sterile PBS at 4 °C for up to 3 days.
3. Polydimethylsiloxane (PDMS) Microstamp Fabrication
NOTE: The fabrication of a silicon master is required prior to start the PDMS microstamp fabrication. This microfabrication of a silicon master can be done by lithographic techniques, which requires specialized equipment and training. Collaborations with a nanofabrication facility are encouraged to fabricate the silicon master. Alternatively, contact a company that fabricates custom-made microstructured silicon masters on demand. It is important to note that the fabrication of the silicon master needs only to be done once. Indeed, microstructured silicon masters can be used indefinitely to produce elastomeric stamps.
- Mix PDMS and the curing agent in a 10:1 ratio in a plastic beaker and mix thoroughly using a pipette for 10 min.
- Degas the PDMS mixture under vacuum to remove air bubbles that were formed during the step 3.1.
- Place the microstructured silicon master in a Petri dish and cast a 10-mm thick layer of degassed PDMS mixture on it without forming bubbles.
- Let cure the PDMS for 2 hr at 60 °C in an oven.
- In a dust-free environment, peel-off the PDMS layer and excise 1 cm2 microstamps with a scalpel.
- Using forceps, place PDMS microstamps pattern-up in a Petri dish.
4. Micropatterning Hydroxy-PAAm Hydrogels
- Place PDMS microstamps in an ethanol/water (50/50) solution and sonicate for 15 min.
- Dry the stamps with a stream of nitrogen flow and place them pattern-up in a UV/Ozone cleaner (λ < 200 nm) for 7 min.
- Under a sterile hood, place a 150-µl drop of a desired protein solution (e.g., 100 µg/ml laminin in PBS or 25 µg/ml fibronectin in PBS) onto the microstructured surface of a 1-cm2 PDMS stamp.
- Optional: Modify the concentration of the protein solution to modulate the cell-ligand density.
- Spread the protein solution across the stamp surface by moving it with a sterile tip of a pipette toward each corner of the stamp.
- Leave the protein solution to adsorb on the PDMS stamp for 60 min under a sterile hood. Turn off lamps to avoid protein damage.
- Under a sterile hood, transfer hydroxy-PAAm coated coverslips (available from step 2.13) into a Petri dish.
- Remove excess PBS from the surface of hydroxy-PAAm substrates with a low nitrogen stream under sterile conditions. Stop the procedure as soon as no evidence of standing water on the gel surface is observed. The gel should not be dried thoroughly at this stage.
- Dry carefully the structured surface of the PDMS stamp with a steady flow of high-purity nitrogen gas.
- Grasp the protein-coated stamp with dressing tissue forceps and place the structured surface in contact with the dried hydrogel surface. Apply brief pressure points with the tip of tweezers on the top of the PDMS stamp to ensure a good contact between stamp microfeatures and the hydrogel surface.
- Leave the PDMS stamp on the hydrogel surface for 1 hr at RT.
- Gently remove PDMS stamps from hydroxy-PAAm hydrogels with dressing tissue forceps and follow the step 4.1 to clean the stamp.
- Wash extensively the stamped hydroxy-PAAm hydrogels by 3 exchanges of PBS (pH = 7.4) in sterile conditions for 10 min per exchange.
- Optional: Additional micropatterns of other ECM proteins can be added to the hydroxy-PAAm surface by following steps 4.5 to 4.11.
- Passivate non-printed zones with a sterile solution of BSA at 5 mg/ml in PBS during one night at 4 °C under a gentle agitation on a rocking plate.
- Wash extensively by 3 exchanges of PBS (pH = 7.4) in sterile conditions for 10 min per exchange. At this stage, stamped hydroxy-PAAm hydrogels can be stored at 4 °C up to one week.
5. Cell Deposition on Micropatterned hydroxy-PAAm Hydrogels
- Incubate coverslips in cell media for 30-45 min prior to plating cells.
- Wash adherent cells cultured in a 75 cm2 culture flask with sterile PBS at 37 °C and detach it with 3 ml of trypsin-EDTA or accutase for 10 min.
- Transfer the desired amount of pre-warmed complete growth medium appropriate for your cell line into the flask containing the detached cells and centrifuge the cell suspension for 3 min at 650 x g.
- Remove the supernatant with a micropipette and resuspend the cells in complete culture medium at 15-20,000 cells/ml.
- Add 4 ml of the cell solution to a micropatterned coverslip (obtained from Step 5.1) and place the cell-covered coverslip in a culture incubator at 37 °C and 5% humidity for 1-2 hr.
- Aspirate gently unattached cells and replace culture medium. Return the attached cells to the incubator and let them spread fully (3-6 hr, depending on the cell type).
Representative Results
Figure 1A presents the co-polymerization of acrylamide (AAm) and bisacrylamide (bis-AAm) with N-hydroxyethylacrylamide (HEA) monomers containing a primary hydroxyl formed by random radical polymerization a hydrophilic network of polyacrylamide with embedded hydroxyl groups (hydroxy-PAAm). In this protocol, a weight 65 mg of HEA must be diluted in a volume of 1 ml of HEPES. Knowing that the density of HEA is roughly equal to one, we assume that we obtain a working volume of 1,065 µl (HEA+HEPES). As presented in Table 1, the total addition of 1,065 µl to 400 µl of AAm and 50 µl of bisAAm leads to a volume of 1,515 µl. Therefore a volume of 3,485 µl of HEPES is needed to adjust the solution to a final volume of 5 ml. As indicated in the step 2.6, the surface of the 22 mm glass coverslip must be activated during 7 min in a UV/Ozone in order to remove it without causing any damages to the hydrogel surface. Indeed, the glass surface becomes more hydrophilic after the UV/Ozone treatment and can be therefore easily removed by immersing the whole system in water. In addition, the UV/Ozone treatment prevents chemical contamination of the 22 mm coverslip, which is in direct contact with the hydrogel surface. This technique permits to obtain a flat circular polyacrylamide surface (22 mm in diameter) bound to a glass coverslip (25 mm in diameter).
As presented in Figure 1B, micropatterns of proteins can be created on the surface of hydroxy-PAAm hydrogels by microcontact printing. In this experimental procedure, UV/Ozone exposures permit to reduce temporarily the intrinsic hydrophobicity of the PDMS stamps by forming silanol groups at the surface. Indeed, the 185 nm line produces ozone from molecular oxygen while the 254 nm line converts the ozone to atomic oxygen. This reactive species attacks the siloxane backbone of PDMS to form oxygen rich SiOx silica-like layer and Si-OH surface groups. The oxidized PDMS surface is known to recover its hydrophobicity in just hours after exposure to air due to the migration of low molecular weight uncrosslinked polymeric chains from the bulk phase to the surface. This strategy helps the spreading of the protein solution on the surface of the PDMS stamp.
One of the key advantages of this method is to independently modulate matrix stiffness (Figure 2A), micropattern geometry (Figure 2B), cell-ligand density (Figure 2C), and protein nature (Figures 3A and 3B). As shown in Figure 2A, hydroxy-PAAm hydrogels show a linear dependence of elastic moduli on cross-linker concentration from few kPa to several dozen kPa, allowing fine reproduction of the rigidity of the cellular microenvironment. Figure 2B presents epifluorescent images of laminin (LM) rectangular micropatterns deposited on hydroxy-PAAm hydrogels of three different stiffnesses (2.5, 15, and 40 kPa), demonstrating clearly the independent tuning of micropattern geometry and matrix stiffness. Figure 2C shows that the cell-ligand density can be modulated by varying the concentration of the protein solution used to incubate PDMS stamps. Figure 3 presents a fluorescence image of collagen lines stamped across laminin and fibronectin lines by using sequential microcontact printings.
It is interesting for the readers to note that various sizes and shapes of micropatterns have been stamped efficiently, regardless the hydrogel stiffness. We have reproduced successfully microfeatures of proteins with sharp edges (e.g., triangles and stars) and straight (e.g., lines), or curved (e.g., circles) shapes, demonstrating that there is no major limitation to the pattern complexity. For single cell experiment, the micropattern surface area was between 400 and 2,500 µm2, the lower value corresponding to the limit of single cell viability. As many other microprinting-based methods, the main limitation of the presented method concerns its spatial resolution. On stiff hydrogels (E > 10 kPa), the spatial resolution is around the micrometer and mainly determined by the resolution of the stamp, whereas on "soft" hydrogels, the limit of resolution becomes partly determined by the surface deformation that may occur during the protein transfer.
The toxicity of hydroxy-PAAm hydrogels was investigated by quantifying the viability of primary endothelial cells plated for 1, 2, and 3 days in culture on homogeneously coated and microprinted soft hydroxy-PAAm hydrogels (Figure 4A). About 80% of HUVECs plated on micropatterned soft hydroxy-PAAm hydrogels maintained their viability for three days, demonstrating the biocompatibility of hydroxy-PAAm hydrogels for cell culture. Interestingly, similar viability was obtained in our laboratory with primary cortical neuron cells. In addition, the primary endothelial cells proliferate more on “stiff” (500 kPa) compared to “soft” (25 kPa) substrates (Figure 4B), indicating that this method provides an appropriate microenvironment for studying cellular proliferation across a wide range of rigidities.
To decouple the effect of substrate stiffness and spreading area on mechanotransduction, primary endothelial cells (HUVECs) were plated on rectangular FN-coated micropatterns (constant area of 1,200 µm2) deposited on hydroxy-PAAm hydrogels of three different stiffnesses (2.5, 8.5, and 25 kPa). Then, HUVECs were stained for actin filaments, vinculin and DNA by following a standard immunocytochemistry procedure, as used for mammalian cells plated on glass substrates. On the softest substrates, HUVECs exhibit low actin fiber density and a rounded nucleus, whereas on stiffer substrates, actin fibers are straighter and thicker and the nucleus deformed (Figure 5). Changes in substrate stiffness at constant spreading area modulate tension and distribution of actin stress fibers, which results in the remodeling of the nucleus. This observation suggests a mechanical coupling between matrix stiffness, cytoskeleton, and nucleus.
| Final AAm/bisAAm ratio | AAm 40% (µl) | Weight of HEA to dissolve in 1 ml HEPES (mg) | bis AAm (2%) (µl) |
HEPES (µl) | Young’s modulus (103 Pa) |
| 0.2/50 0.5/50 1/50 2/50 3/50 |
400 400 400 400 400 |
65 65 65 65 65 |
50 125 250 500 750 |
3,485 3,410 3,285 3,035 2,785 |
~1.4 ~ 3.6 ~ 8.7 ~17.2 ~ 25 |
Table 1. Preparation of hydroxy-PAAm hydrogels with various rigidities. Working solutions to prepare hydroxy-PAAm hydrogels with final stiffness ranging from 1.4 to 25 kPa.

Figure 1. (A) Acrylamide (AAm, in black), N,N’-methylenebisacrylamide (bis-AAm, in blue) and N-hydroxyethylacrylamide (HEA, in red) were mixed together to form hydroxy-PAAm hydrogels. (B) Schematic representation of the different steps of the micropatterning procedure on hydroxy-PAAm hydrogels. A solution of protein is first incubated during 1 hr on the structure face of a microstamp (step #1). The hydrogel surface is gently dried with a nitrogen flow (step #2). The microstamp is then placed in formal contact with the hydroxy-PAAm surface during 1 hr (step #3). After successive washes, the microprinted surface is passivated with a sterile BSA solution deposited O/N at 4 °C (step #5) to block non-printed areas. Finally, the microprinted hydroxy-PAAm surface is washed several times with sterile PBS and is ready for cell seeding (step #6). Please click here to view a larger version of this figure.
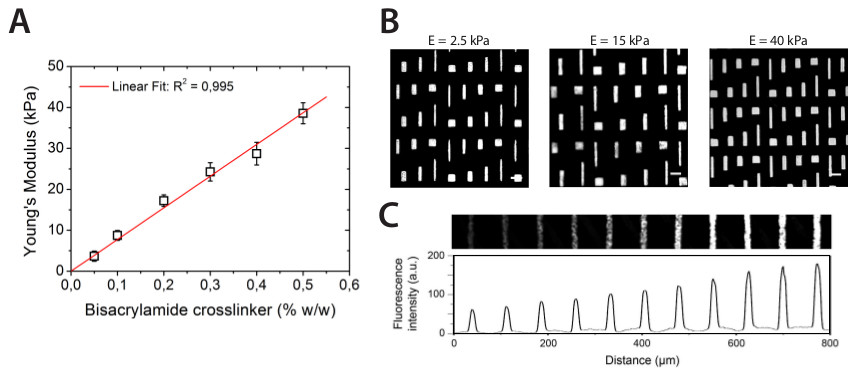
Figure 2. (A) The evolution of the stiffness of hydroxy-PAAm hydrogels is linearly correlated with the amount of bis-AAm cross-linker (the red line is a linear regression with R2=0,995). (B) Fluorescence images of rectangular laminin micro-features stamped on hydroxy-PAAm hydrogels of different stiffnesses (E = 2.5, 15, and 40 kPa). Scale bars correspond to (A) 65 µm, (B) 80 µm, and (C) 50 µm. (C) Parallel lines of 15 µm width form a LM gradient on a 25 kPa hydroxy-PAAm hydrogel, as indicated by the evolution of the fluorescence intensity profile. Please click here to view a larger version of this figure.

Figure 3. (A) Immunofluorescence image of Fibronectin (in red) and Laminin (in green) stripes crossed at 90°. (B) Multilabeling of Fibronectin (in red), Laminin (in green) and collagen (in blue) stripes microprinted subsequently on a 25 kPa hydrogel PAAm substrate. Scale bars correspond to 30 µm. Please click here to view a larger version of this figure.
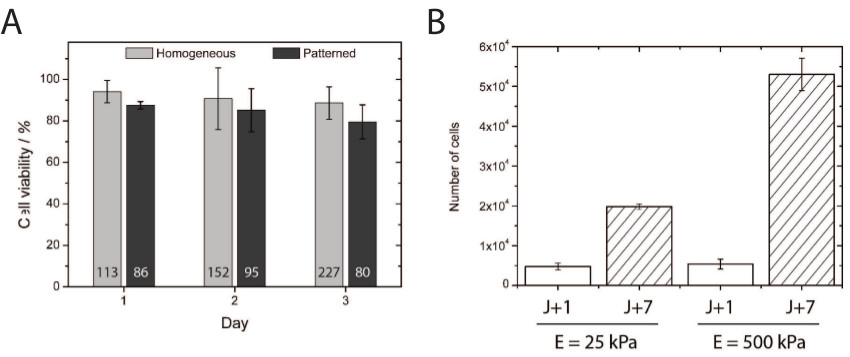
Figure 4. (A) The quantification of cell viability demonstrated excellent cell survival at 24, 48, and 72 hr after being plated on homogeneously-coated and micropatterned hydroxy-PAAm surfaces. The number indicated at the bottom of each bar corresponds to the total number of cells counted for the viability assay. (B) Quantification of HUVECs proliferation on 25 and 500 kPa homogeneously-coated substrates after 1 (white bars) and 7 (hashed bars) days of culture. Please click here to view a larger version of this figure.
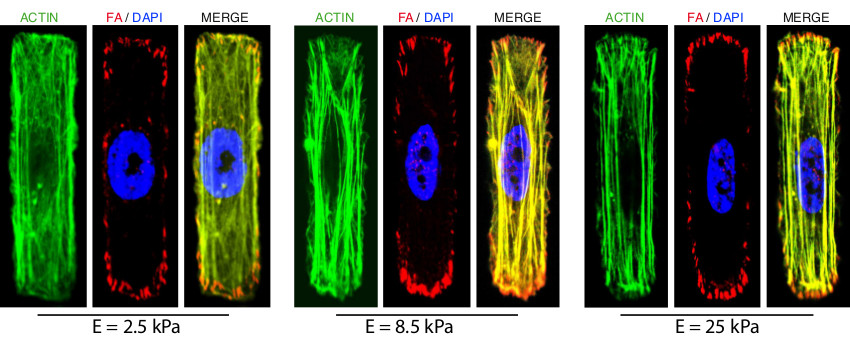
Figure 5. Fluorescent images of immunostained primary endothelial cells cultured on rectangular FN-coated micropatterns, which were deposited on hydroxy-PAAm hydrogels of 2.5 kPa, 8.5 kPa, and 25 kPa (from left to right). The actin stress fibers (in green) were stained with Alexa Fluor 488 phalloidin, focal adhesions (in red) were stained with a mouse polyclonal primary antibody and labeled with a red-fluorescent IgG secondary antibody and the DNA was stained in blue with DAPI. The scale bars correspond to 17 µm. Please click here to view a larger version of this figure.
Discussion
Many in vitro observations in modern cell biology have been performed on rigid glass coverslips, often coated with a thin layer of ECM proteins or synthetic peptides containing the RGD sequence. However, such basic culture substrates do not recapitulate the whole physicochemical complexity of the ECM and thus do not provide an accurate model for studying cellular mechanotransduction processes. To tackle this problem, we propose a simple alternative to functionalize two-dimensional hydrogels with any desired amount and nature of ECM proteins. This method allows independent control of important mechanotransduction cues, such as matrix stiffness, cellular morphology and confinement or cell-ligand density. In addition, hydroxy-PAAm hydrogels overcome one of the greatest limitations of current functionalization methods, which is the inability to immobilize and to control the spatial distribution of more than one ECM protein. Due to the high affinity of hydroxy-PAAm hydrogels for biomolecules, one can control the spatial distribution of various ECM proteins on the same surface, regardless the matrix stiffness, by using sequential microprintings, which do not require any supplementary equipment.
We propose a novel procedure based on the incorporation of hydroxyl groups within acrylamide hydrogels to surmount their intrinsically non-adhesive properties with minimal requirements in cost or expertise. In comparison with existing techniques for patterning proteins on hydrogels, the hydroxy-PAAm method avoids the use of harsh toxic chemicals (e.g., hydrazine hydrate), expensive photoreactive crosslinkers (e.g., Sulfo-Sanpah) and the dependence on UV lamp power and positioning to functionalize soft hydrogels. Hydroxy-PAAm hydrogels remain stable and active for several weeks, can be mass produced, easily microprinted and their high affinity for biomolecules allows to investigate synergistic effects of conjugated ECM proteins on cellular functions. In addition, hydroxy-PAAm hydrogels can be used to quantitatively investigate the amount of contractile forces exerted by cells on their surrounding microenvironment. Indeed, we have shown previously1,20 that fluorescent beads can be easily embedded in hydroxy-PAAm hydrogels, in order to use cell traction force microscopy (TFM) to measure the displacement field and estimate the resulting traction field exerted by eukaryotic cells plated on two-dimensional micropatterns.
As many other microprinting-based methods, the main limitation of hydroxy-PAAm hydrogels concerns the spatial resolution of protein microfeatures. On stiff hydrogels (E > 10 kPa), the spatial resolution is around a micrometer and mainly determined by the resolution of the stamp, depending on the lithography technique used to fabricate the silicon mold. On softer hydrogels, the spatial resolution becomes partly determined by the surface deformation during the protein transfer, which becomes the principal limitation to reach micrometer spatial resolution. The approach presented here is currently limited to two-dimensional substrates. However this method is highly scalable and further studies are currently underway in our laboratory to extend this method to three-dimensional structures.
We hope that hydroxy-PAAm hydrogels may help to decipher key mechanisms of complex cellular and tissue processes that are intimately related to the physicochemical properties of the cell microenvironment21,22, including embryonic development, inflammatory responses, wound healing, neurite outgrowth, adult tissue homeostasis, and the pathogenesis of diseases such as fibrosis and cancer.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the Belgian National Foundation for Scientific Research (F.R.S.-FNRS) through “MIS Confocal Microscopy”, “Crédit aux Chercheurs” grants and the “Nanomotility FRFC project” (no. 2.4622.11). T.G. doctoral fellowship is supported by the Foundation for Training in Industrial and Agricultural Research (FRIA). The authors gratefully acknowledge Sylvain Desprez for mechanical characterization and Géraldine Circelli for confocal imaging.
Materials
| UV/Ozone Photoreactor | Ultra-Violet Products | Model PR-100 | |
| Rocking plate | IKAcWerke | Model KS 130 Basic | |
| Vortexer | Scientific Industries | Model Vortex Genie2 | |
| Vacuum degassing chamber | Applied Vacuum Engineering | DP- 8-KIT | |
| Parafilm | Sigma-Aldrich | P7793-1EA | |
| Stainless steel forceps with fine tip | Sigma-Aldrich | Z225304-1EA | |
| Dressing tissue forceps | Sigma-Aldrich | F4392-1EA | |
| Petri dishes in polystyrene | Sigma-Aldrich | P5731-500EA | |
| Aluminium foil, thickness 0.5 mm | Sigma-Aldrich | 266574-3.4G | |
| Isopore membrane filter (0,2 µm pore size) | Millipore | GTTP Filter code | |
| Round glass coverslip (22 mm diameter) | Neuvitro | GG-22 | |
| Round glass coverslip (25 mm diameter) | Neuvitro | GG-25 | |
| Variable volume micropipette | Sigma-Aldrich | Z114820 | |
| Protein microcentrifuge tubes | Sigma-Aldrich | Z666505-100EA | |
| Scalpel handles | Sigma-Aldrich | S2896-1EA | |
| Scalpel blades | Sigma-Aldrich | S2771-100EA | |
| Cell culture flasks (75 cm2) | Sigma-Aldrich | CLS430641 | |
| Ultrasonic bath tray, solid (stainless steel) | Sigma-Aldrich | Z613983-1EA | |
| Name of the Reagent | Company | Catalogue Number | Comments |
| Polydimethylsiloxane | Dow Corning | Sylgard 184 silicone elastomer kit | |
| Acrylamide (powder) | Sigma-Aldrich | A3553 | |
| N,N’-Methylenebis(acrylamide) | Sigma-Aldrich | 146072 | |
| N-Hydroxyethylacrylamide | Sigma-Aldrich | 697931 | |
| N,N,N’,N’-Tetramethylethylenediamine | Sigma-Aldrich | T9281 | |
| Amonium PerSulfate (APS) | Sigma-Aldrich | A3678 | |
| 3-(Trimetoxysilyl)propyle acrylate | Sigma-Aldrich | 1805 | |
| Human Plasma Fibronectin | Millipore | FC010 | |
| Laminin from EHS | Sigma-Aldrich | L2020 | |
| Sodium hydroxyde | Sigma-Aldrich | 221465-25G | |
| Double-distilled water (ddH2O) | |||
| Endothelial cell growth medium | Cells Applications | 211K-500 | |
| Human Umbilical Vein Endothelial Cells (HUVEC) | Invitrogen | C-003-5C | |
| Accutase | PAA laboratories | L11-007 | |
| HEPES buffer solution 1M in H20 | Sigma-Aldrich | 83264-500ML-F | |
| Antibiotics-antimycotics | PAA laboratories | P11-002 | |
| Phosphate Buffer Saline solution | PAA laboratories | H15-002 | |
| Alexa Fluor 488 Phaloidin | Molecular Probes | A12379 | |
| Anti-vinculin antibody produced in mouse | Sigma-Aldrich | V9131 | |
| Goat anti-mouse antibody-tetramethylrhodamine | Molecular Probes | T-2762 | |
| Anti-Fibronectin | Sigma-Aldrich | F3648 | |
| (rabbit) | |||
| Streptavidin | Sigma-Aldrich | 41469 | |
| Anti-Laminin antibody | Sigma-Aldrich | L9393 | |
| (rabbit) | |||
| Anti-rabbit IgG-FITC | Sigma-Aldrich | F7512 | |
| Trypsin-EDTA solution | Sigma-Aldrich | T3924-100ML | |
| Absolute ethanol | Sigma-Aldrich | 459844-2.5L |
References
- Grevesse, T., Versaevel, M., Circelli, G., Desprez, S., Gabriele, S. A simple route to functionalize polyacrylamide gels for the independent tuning of mechanotransduction cues. Lab Chip. 13 (5), 777-780 (2013).
- Gabriele, S., Benoliel, A. M., Bongrand, P., Théodoly, O. Microfluidic investigation reveals distinct roles for actin cytoskeleton and myosin II activity in capillary leukocyte trafficking. Biophys. J. 96 (10), 4308-4318 (2009).
- Atmanli, A., Domian, I. J. Generation of aligned functional myocardial tissue through microcontact printing. J. Vis. Exp. (73), e50288 (2013).
- Gabriele, S., Versaevel, M., Preira, P., Théodoly, O. A simple microfluidic method to select, isolate, and manipulate single-cells in mechanical and biochemical assays. Lab Chip. 10 (11), 1459-1467 (2010).
- Le Berre, M., Aubertin, J., Piel, M. Fine control of nuclear confinement identifies a threshold deformation leading to lamina rupture and induction of specific genes. Integr. Biol. 4 (11), 1406-1414 (2012).
- Rundqvist, J., et al. High fidelity functional patterns of an extracellular matrix protein by electron beam-based inactivation. J. Am. Chem. Soc. 129 (59), 59-67 (2006).
- Sorribas, H., Padeste, C., Tiefenauer, L. Photolithographic generation of proteins micropatterns for neuron culture applications. Biomaterials. 23 (3), 893-900 (2002).
- Holden, M. A., Cremer, P. S. Light activated patterning of dye-labeled molecules on surfaces. J. Am. Chem. Soc. 125 (27), 8074-8075 (2003).
- Cheng, Q., Li, S., Komvopoulos, K. Plasma-assisted surface chemical patterning for single-cell culture. Biomaterials. 30 (25), 4203-4210 (2009).
- Kumar, A., Whitesides, G. M. Features of gold having micrometer to centimeter dimensions can be formed through a combination of stamping with an elastomeric stamp and an alkanethiol “ink” followed by chemical etching. Appl. Phys. Lett. 63 (14), 2002-2004 (1993).
- Tseng, Q., et al. New micropatterning method of soft substrates reveals that different tumorigenic signals can promote or reduce cell contraction levels. Lab Chip. 11, 2231-2240 (2011).
- Palchesko, R. N., Zhang, L., Sun, Y., Feinberg, A. W. Development of Polydimethylsiloxane Substrates with Tunable Elastic Modulus to Study Cell Mechanobiology in Muscle and Nerve. PLoS ONE. 7, e51499 (2012).
- Herrick, W. G., Nguyen, T. V., Sleiman, M., McRae, S., Emrick, T. S., Peyton, S. R. PEG-Phosphorylcholine hydrogels as tunable and versatile platforms for mechanobiology. Biomacromolecules. 14, 2294-2304 (2013).
- Versaevel, M., Grevesse, T., Riaz, M., Lantoine, J., Gabriele, S. Micropatterning hydroxy-PAAm hydrogels and sylgard 184 silicone elastomers with tunable elastic moduli. Methods in Cell Biology. 121, 33-48 (2014).
- Kandow, C. E., Georges, P. C., Janmey, P. A., Beningo, K. A. Polyacrylamide hydrogels for cell mechanics: steps towards optimization and alternative uses. Methods Cell Biol. 83, 29-46 (2007).
- Hemphill, M. A., et al. A possible role for integrin signaling in diffuse axonal injury. PLoS ONE. 6 (7), e22899 (2011).
- Damljanovic, V., Lagerholm, B. C., Jacobson, K. Bulk and micropatterned conjugation of extracellular matrix proteins to characterized polyacrylamide substrates for cell mechanotransduction assays. BioTechniques. 39 (6), 847-851 (2005).
- Hynd, M. R., Frampton, J. P., Dowell-Mesfin, N., Turner, J. N., Shain, W. Directed cell growth on protein-functionalized hydrogel surfaces. Journal of Neuroscience Methods. 162, 255-263 (2007).
- Tseng, Q., et al. new micropatterning method of soft substrates reveals that different tumorigenic signals can promote or reduce cell contraction levels. Lab Chip. 11 (13), 2231-2240 (2011).
- Versaevel, M., Grevesse, T., Gabriele, S. Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nat. Commun. 3, 671 (2012).
- Versaevel, M., Grevesse, T., Riaz, M., Gabriele, S. Cell Confinement: Putting the squeeze on the nucleus. Soft Matter. 9, 6665-6676 (2013).
- Trappman, B., Chen, C. S. How cells sense extracellular matrix stiffness: a material’s perspective. Current Opinion in Biotechnology. 24, 1-6 (2013).

