Culturas puras e semeadura por esgotamento: isolamento de colônias bacterianas únicas de uma amostra mista
English
Share
Overview
Fonte: Tilde Andersson1, Rolf Lood1
1 Departamento de Ciências Clínicas Lund, Divisão de Medicina de Infecção, Centro Biomédico, Universidade de Lund, 221 00 Lund, Suécia
Aparentemente impossível de determinar, a biodiversidade microbiana é verdadeiramente surpreendente com uma estimativa de um trilhão de espécies coexistindo (1,2). Embora climas particularmente severos, como o ambiente ácido do estômago humano (3) ou os lagos subglaciais da Antártida (4), possam ser dominados por uma espécie específica, as bactérias são tipicamente encontradas em culturas mistas. Como cada cepa pode influenciar o crescimento de outra (5), a capacidade de separar e cultivar colônias “puras” (consistindo apenas de um tipo) tornou-se essencial em ambientes clínicos e acadêmicos. Culturas puras permitem mais exames genéticos (6) e proteômicos (7), análise da pureza amostral e, talvez mais notável, a identificação e caracterização de agentes infecciosos a partir de amostras clínicas.
As bactérias têm uma ampla gama de requisitos de crescimento e existem inúmeros tipos de mídia de nutrientes projetadas para sustentar tanto as espécies não exigentes quanto as meticulosas (8). A mídia de crescimento pode ser preparada na forma líquida (como um caldo) ou em uma forma sólida tipicamente baseada em ágar (um agente de gelling derivado de algas vermelhas). Considerando que a inoculação direta no caldo traz o risco de gerar uma população bacteriana geneticamente diversa ou mesmo mista, chapeamento e re-streaking cria uma cultura mais pura onde cada célula tem uma composição genética altamente semelhante. A técnica da placa de raia baseia-se na diluição progressiva de uma amostra(Figura 1),com o objetivo de separar células individuais umas das outras. Qualquer célula viável (doravante referida como uma unidade formadora de colônias, CFU) sustentada pela mídia e ambiente designado pode posteriormente encontrar uma colônia isolada de células filhas através de fissão binária. Apesar das rápidas taxas de mutação dentro das comunidades bacterianas, esse grupo celular é geralmente considerado clonal. A colheita e re-streaking dessa população, consequentemente, garante que o trabalho subsequente envolva apenas um único tipo de bactéria.
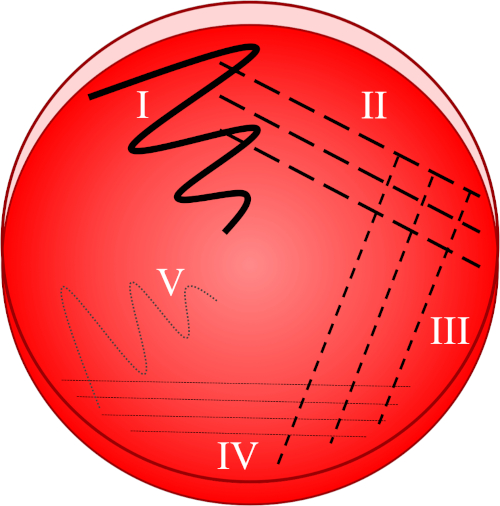
Figura 1: Uma placa de raia é baseada na diluição progressiva da amostra original. I) O inóculo é inicialmente disperso usando um movimento em zig-zag, criando uma área com uma população bacteriana relativamente densa. II-IV) As listras são extraídas da área anterior, usando um laço de inoculação estéril cada vez, até que o quarto quadrante seja atingido. V) Um movimento final em zigue-zague direcionado para o meio da placa forma uma região onde o inóculo foi marcadamente diluído, permitindo que as colônias apareçam separadas umas das outras.
A técnica da placa de raia também pode ser combinada com o uso de mídias seletivas e/ou diferenciais. Um meio seletivo inibirá o crescimento de certos organismos ( porexemplo, através da adição de antibióticos), enquanto um meio diferencial só ajudará a distinguir um do outro (por exemplo, através da hemolise em placas de ágar sanguíneo).
Por trás de todo o trabalho em microbiologia está o uso de técnicas assépticas (estéreis). Toda cultura bacteriana deve ser considerada potencialmente patogênica, pois há risco de crescimento não intencional de cepas traiçoeiras, formação de aerossol e contaminação de equipamentos/pessoal. Para minimizar esses riscos, todas as mídias, plásticos, metal e vidro são tipicamente esterilizados através de autoclaving antes e depois do uso, submetendo-os a vapor saturado de alta pressão em torno de 121°C que efetivamente elimina quaisquer células persistentes. O espaço de trabalho é geralmente desinfetado usando etanol antes e depois do uso. Jaleco e luvas são sempre usados durante o trabalho com agentes infecciosos.
Procedure
Results
The initial streak-plate may contain colonies originating from cells with different genetic makeup or (depending on sample purity) from different bacterial species (Figure 2A).
Through subsequent isolation of a single colony, where all units are derived from a common mother-cell, the second streaking procedure generates a relatively clonal bacterial population, suitable for further characterization or inoculation into broth (Figure 2B).

Figure 2: A pure culture can be generated from a mixed sample through isolation of a single, secluded colony. A) Growth of a single bacterial cell (CFU) generated a clonal colony, separated from those of other species and strains. This CFU was used for subsequent streaking onto a new plate B) A second plate, where the bacterial population consists solely of cells derived from the initial CFU.
Applications and Summary
The ability to obtain and cultivate a pure bacterial colony is essential, both in clinical and academic settings. Streak plating enables the isolation of a relatively clonal cell population, originating from a shared CFU, that may be of particular interest during diagnosis or for additional characterization of the isolate. A sample is streaked onto a suitable agar-based nutrient medium and incubated until colonies become visible. An isolated colony is subsequently harvested and re-streaked onto a second plate.
References
- The Human Microbiome Project C. Structure, Function and Diversity of the Healthy Human Microbiome. Nature. 486:207-214. (2012)
- Locey KJ, Lennon JT. Scaling laws predict global microbial diversity. Proceedings of the National Academy of Sciences. 113 (21) 5970-5975 (2016)
- Skouloubris S, Thiberge JM, Labigne A, De Reuse H. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infection and Immunity. 66:4517-21. (1998)
- Mikucki JA, Auken E, Tulaczyk S, Virginia RA, Schamper C, Sørensen KI, Doran PT, Dugan H, Foley N. Deep groundwater and potential subsurface habitats beneath an Antarctic dry valley. Nature Communications. 6:6831. (2015)
- Mullineaux-Sanders C, Suez J, Elinav E, Frankel G. Sieving through gut models of colonization resistance. Nature Microbiology. 3:132-140. (2018)
- Fournier PE, Drancourt M, Raoult D. Bacterial genome sequencing and its use in infectious diseases. Lancet Infectious Diseases. 7:711-23 (2007)
- Yao Z, Li W, Lin Y, Wu Q, Yu F, Lin W, Lin X. Proteomic Analysis Reveals That Metabolic Flows Affect the Susceptibility of Aeromonas hydrophila to Antibiotics. Scientific Reports. 6:39413 (2016)
- Medina D, Walke JB, Gajewski Z, Becker MH, Swartwout MC, Belden LK. Culture Media and Individual Hosts Affect the Recovery of Culturable Bacterial Diversity from Amphibian Skin. Frontiers in Microbiology. 8:1574 (2017)
Transcript
On a Petri dish, if a single bacterium undergoes multiple rounds of asexual reproduction, it will lead to the formation of a clonal colony. However, obtaining a single bacterium from a mixed sample, such as a soil suspension, can be difficult. If one loopful of this heterogeneous culture is taken, it can contain as many as one trillion individual bacteria. To spread this many bacteria out onto the surface of an agar plate and obtain a single colony, even using a zig-zag pattern, the loop would need to be dragged continuously over the surface of enough plates set side-by-side to encircle the entirety of Liberty Island. Obviously, scientists do not really use that many plates. Instead, they use a technique called streak plating.
The streak plate technique is based on progressive dilution of a bacterial sample, and it is performed over the solid media surface of a single Petri dish. To begin, the media surface is visually divided into five sections by assigning four fragments of the circumference as the first four sections, and the plate’s center as the fifth. This will effectively create five media plates out of a single Petri dish. Next, using a loopful of desired inoculum, the first section is streaked using a zig-zag pattern. Then, either a new disposable loop is used, or in the case of a wire loop, it is sterilized with a Bunsen burner, flaming it until it is red hot along the length of the wire. This use of a new loop, or flame sterilize loop, removes any remaining bacterial cells, assisting in the dilution of the bacteria. The hot loop is then cooled in the air for a few seconds before being dragged through the first section to create three to four separate lines, each carrying only a fraction of bacteria into the second section. The remaining sections are streaked in the same manner, using a sterile loop each time, and a single pass through the previous streak.
Using this cycle of streaking and sterilizing, the bacterial concentration in every subsequent section should be diluted so that the final section contains only a few discretely located bacteria. Upon incubation, these discrete bacteria multiply to produce isolated clonal colonies of daughter cells, which are referred to as Colony Forming Units, or CFUs. These can be harvested and re-streaked to ensure that subsequent work involves only a single bacterial type, referred to as a pure culture. As well as isolating single colonies from a mixed-bacterial culture, the streak plating technique is also used to select media-specific strains, determine bacterial colony morphology, or identify different bacterial species. In this video, we will demonstrate how to isolate single-bacterial colonies from a mixed-bacterial sample suspension via streak plating technique.
To begin, put on laboratory gloves and a lab coat. Next, sterilize the workspace using 70% ethanol. Next, select a suitable medium that will sustain the utilized bacterial species or strain and begin preparing the media. Here, common LB agar is prepared by weighing out ten grams of pre-formulated, powdered media and 7.5 grams of agar. Add the weighed, dried components to a glass bottle which is able to hold twice the final volume to avoid overflow. Then, add 500 milliliters of water to the bottle, and cap it semi-tightly. Sterilize the media by placing the bottle in an autoclave set to 121 degrees Celsius for twenty minutes. After completion, use heat-proof gloves or a hot pad to remove the media from the machine and then immediately twist the bottle cap to close it tightly.
For the same-day use, let the media cool down by placing the bottle into a water bath heated to approximately 45 degrees Celsius, to preserve the media in a liquid state. Alternatively, the media can be left at room temperature to store at solid state. When needed, microwave the bottle with the lid slightly open to melt the media, and allow the media to cool using a 45 degree Celsius water bath.
Next, take a sleeve of sterile Petri dishes, and with a permanent marker, label them with the investigator and media names as well as the date. Then, transfer the required volume of media into a sterile vessel, and add antibiotics or other sensitive components if necessary. Here, 50 milliliters of media is mixed with 100 microliters of Kanamycin for a final concentration of 25 micrograms per milliliter. Swirl the tube to ensure even distribution of the added components throughout the media. Slowly, so as to avoid bubble formation, pour 20 to 25 milliliters of approximately 45 degree Celsius culture medium into each of the plates. If bubbles or foam appear, swiftly remove using a regular pipette and a sterile tip. Then, immediately replace all lids to prevent contamination. Allow the agar to solidify at room temperature for at least two hours or overnight. Once solidified, store the culture plates upside down at four degrees Celsius to minimize condensation on the medium’s surface.
To streak the culture of choice, first take a clean culture plate and remove the lid. Working quickly, submerge a disposable, sterile loop into the desired inoculum and then immediately swab the loop over the first quadrant of the plate using a zig-zag motion. Replace the lid of the dish, discard the used inoculation loop, and then select a new sterile loop. Using the new loop, make three to four strokes crossing the original swab line radiating from the first quadrant, which should contain a relatively dense bacteria population into the second quadrant. Close the lid once more, and discard the loop. With a new loop, repeat this action again, but this time streaking from the second into the third quadrant. Then, with a new loop again, make another streak from the third into the fourth section of the plate. Finally, with a fresh loop, make one last stroke in a zig-zag pattern from the fourth quadrant towards the center of the plate. The bacterial prevalence will be lower in this area, ideally allowing individual colonies to be established from a single viable mother cell.
Replace the plate lid, and if appropriate for the bacterial species, seal the plate with para film to prevent airflow. Turn the culture plate upside down to prevent condensation drips, and then place at a suitable temperature for growth. Here, an incubator is set to 37 degrees Celsius. Allow the plate to incubate until bacterial colonies are visible. To generate a clonal bacterial population, select one discrete colony from this plate. Now, with the sterile loop, touch the target colony, and as before, make a streak in the first quadrant of a new plate. Continue to alternately sterilize the loop and streak the remaining quadrants of the plate as previously demonstrated, ending with the zig-zag to the center. Close the plate, and place it to incubate until discrete colonies form. Once these colonies are grown, they will typically represent pure clonal strains.
The initial streak plate may contain colonies originating from cells from different bacterial species or cells with different genetic makeup, depending upon the sample purity. Through subsequent isolation of a single colony, where all units are derived from a common mother cell, the second streaking procedure generates a relatively clonal bacterial population, suitable for further characterization or inoculation into broth.
