结合:将阿霉素耐药性从捐赠者转移到受体大肠杆菌的方法
English
Share
Overview
资料来源:亚历山大·金1,托尼娅·科尔皮茨1
1波士顿大学医学院微生物学系,国家新兴感染疾病实验室,波士顿,马萨诸塞州
1946年,莱德伯格和塔图姆首次发现,结合是细菌之间的一种水平基因转移形式,依赖于两个细菌细胞之间的直接物理接触(1)。与其他形式的基因转移(如转化或转导)不同,结合是一个自然发生的过程,DNA以单向的方式从供体细胞分泌到受体细胞。这种方向性和这一过程增加细菌遗传多样性的能力,使结合作为细菌”交配”的一种形式的声誉,这被认为极大地促进了最近抗生素耐药性的上升细菌(2,3)。通过使用选择性压力,例如使用抗生素,结合纵用于实验室设置,使其成为细菌之间水平基因转移的有力工具,在某些情况下,从细菌到酵母、植物和动物单元格 (4)。除了在实验室中的应用,通过结合转移真核酸细菌基因是DNA转移的一个令人兴奋的途径,具有多种可能的生物技术应用和自然发生的影响(5)。
结合被认为是通过”两步机制”(6)工作。首先,在转移任何DNA之前,供体细胞必须与受体进行细胞对细胞的直接接触。这个过程在革兰氏阴性细菌中具有最佳特征,其中研究最多的是大肠杆菌。细胞与细胞接触是通过在捐赠者身上存在一个复杂的细胞外细丝网络而建立的,这种结合因子由称为F(生育)因子(7,8)的可转移基因编码。除了在供体和受体之间建立接触外,几种蛋白质通过性皮球输送到受体细胞质,在两个细胞之间形成IV型分泌系统(T4SS)导管,这是第二步的必要结构。共和,DNA转移(6)。通过将性皮鲁斯的这种功能与DNA的滚动循环复制相结合,供体细胞能够通过”芽和泵”模型(6)将DNA以可转位元素的形式(如质粒或转位子)转移给受体。在这种情况下,”射击”是通过T4SS将具有链接DNA的先导蛋白输送到受体细胞,而”泵送”是DNA向受体的主动传输,这个过程依赖于T4SS,并通过耦合蛋白(6)催化。这个过程中使用的机械由转移序列(oriT)的起源组成,必须由CIS和转基因中的DNA提供,该DNA编码松弛酶、配偶配对形成复合体和IV型耦合蛋白,以及可以存在于cis或转(9) 中。这种松弛酶在oriT序列中分裂尼克位点,并共价附着在转移链的5’端,以产生松弛体,一种单链DNA-松弛酶复合物与其他辅助蛋白(9)。一旦形成,松弛体通过IV型耦合蛋白连接到交配对形成复合体,它允许SsDNA-松弛酶复合物通过T4SS(10)转移到受体细胞中。一旦进入受体的细胞质,DNA可以集成到受体基因组中,或者以质粒的形式单独存在,其中任何一种都允许其基因的表达。
在本实验中,使用广泛使用的偶联供体菌株大肠杆菌WM3064将对受体大肠杆菌J53的抗中青霉素的基因编码转移。 虽然革兰氏阴性细菌的两种菌株对四环素具有抗药性,但只有供体菌株WM3064具有氨基青霉素抗性基因,编码为pWD2-oriT穿梭载体中,对二氨基甲酸(DAP)(DAP)具有辅助营养性。该实验包括两个主要步骤,一是制备供体和受体菌株,然后是通过结合将安培林抗性基因从供体转移到受体(图1)。
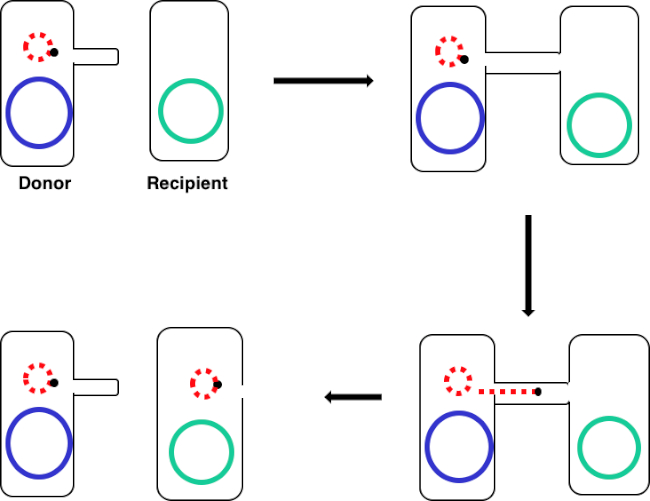
图1:共通示意图。此原理图显示了质粒(仅可转位DNA元素的一个示例)从供体细胞使用结合从受体细胞成功转移。当供体细胞通过性皮鲁斯与受体细胞接触时,质粒通过滚动循环复制复制,通过连接两个细胞的多蛋白复合体移动,并在受体细胞中形成新的全长质粒。
通过孵育供体和受体细胞的混合物,然后在四环素和DAP的存在下连续电镀这些细胞,从而成功转移了阿霉素抗性基因。在四环素和安平存在的情况下,从这种混合物中生长的连续电镀细胞,由于缺少DAP和可能尚未获得环青素抗基因的任何受体细胞,去除了所有供体细胞,产生严格受体J53菌株获得对青霉素的细菌(图2)。一旦进行,通过PCR证实了青霉素抗性基因的成功转移。由于结合成功,大肠杆菌的J53菌株含有pWD2-oriT,对阿霉素具有抗药性,PCR可检测到这种抗性的基因编码。然而,如果不成功,将没有检测到阿霉素耐药性基因,而阿霉素仍将作为对抗J53菌株的有效抗生素。
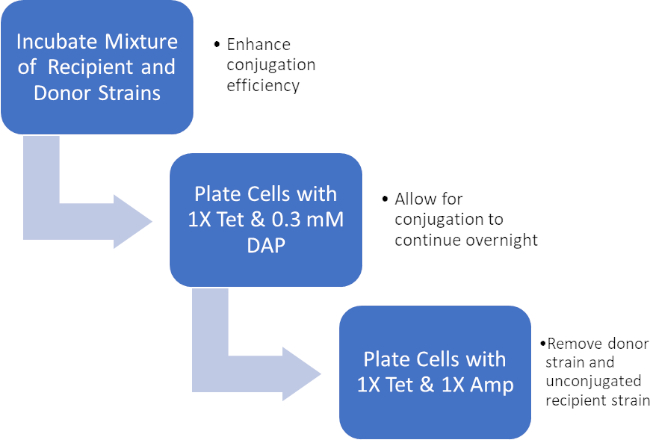
图2:协议示意图。此原理图显示了所呈现协议的概述。
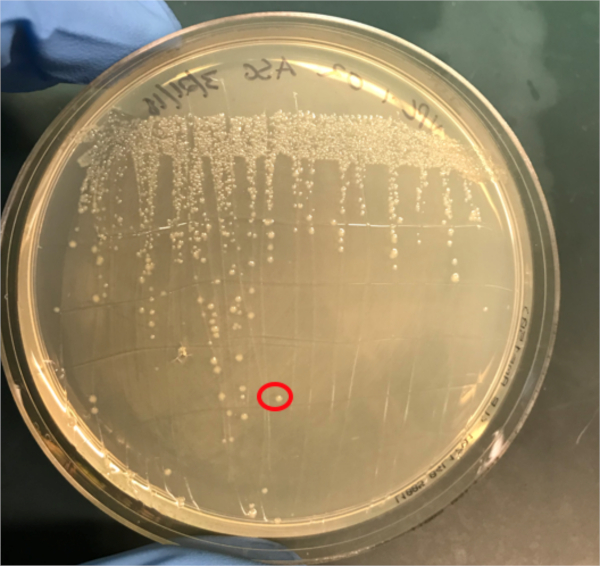
图3A:PCR成功结合的确认。A) 在琼脂板上条纹的偶联和阴性对照样本的冷冻库,并选择一个菌群(红色)进行DNA分离。
Procedure
Results
If conjugation was successful, a 500 base-pair sized band PCR product will be observed in the well in which PCR reaction 1 was loaded (Well #2 in Figure 3B), while no bands will be observed in the well in which PCR reaction 3 was loaded (Well #4 in Figure 3B). The presence of this band confirms the successful transfer of the ampicillin resistance gene, thereby conferring ampicillin resistance to the J53 strain of E. coli.

Figure 3B: The confirmation of successful conjugation by PCR. B) PCR analysis was done using DNA isolated from the select colony. The contents of each well are as follows: 1) DNA ladder, 2) Conjugation DNA and ampicillin primers, 3) Conjugation DNA and housekeeping primers, 4) Negative control DNA and ampicillin primers, 5) Negative control DNA and housekeeping primers, 6) No DNA and ampicillin primers, and 7) No DNA and negative control primers. The presence of a ~ 500 base-pair band PCR product from PCR reaction 1 (well 2), and the lack of this product from PCR reaction 3 (well 4), confirms successful conjugation.
Applications and Summary
Conjugation is a naturally occurring process of horizontal gene transfer that relies on the direct cell-to-cell contact of a donor cell and a recipient cell. This process is shared among all kinds of bacteria and has been instrumental in bacterial evolution, most notably antibiotic resistance. In the lab, conjugation can be used as an effective method of gene transfer that is much less disruptive when compared to other techniques. Outside of the laboratory, the ability to transfer DNA from bacteria to eukaryotes via conjugation offers an exciting new avenue of gene therapy and understanding the implications of these naturally occurring gene transfers, for example the relationship between bacterial infection and cancer, is a rapidly emerging area of research.
References
- Lederberg J, Tatum, E.L. Gene recombination in Escherichia coli Nature. 1946;158:558.
- Holmes R.K. J, M.G. Genetics: Exchange of Genetic Information. 4th Edition ed. Baron S, editor. Galveston, TX: University of Texas Medical Branch at Galveston; 1996.
- Cruz F, Davies, J. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends in Microbiology. 2000;8:128-33.
- Llosa M, Cruz, F. Bacterial conjugation: a potential tool for genomic engineering. Ressearch in Microbiology. 2005;156:1-6.
- Lacroix B, Citovsky, V. Transfer of DNA from Bacteria to Eukaryotes. mBio. 2016;7(4):1-9.
- Llosa M, et al. Bacterial conjugation: a two-step mechanism for
- DNA transport. Molecular Microbiology. 2002;45:1-8.
- Grohmann E, Muth, G., Espinosa, M. Conjugative Plasmid Transfer in Gram-Positive Bacteria. Microbiology and Molecular Biology Reviews. 2003;67:277-301.
- Firth N, Ippen-Ihler, K, Skurray, RA. Structure and function of the F factor and mechanism of conjugation. Escherichia coli and salmonella: cellular and molecular biology. 1996;2:2377-401.
- Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EPC, De La Cruz F. Mobility of Plasmids. Microbiology and Molecular Biology Reviews. 2010;74(3):434-52.
- Cascales E. Definition of a Bacterial Type IV Secretion Pathway for a DNA Substrate. 2004;304(5674):1170-3.
- Wang P, Yu Z, Li B, Cai X, Zeng Z, Chen X, et al. Development of an efficient conjugation-based genetic manipulation system for Pseudoalteromonas. Microbial Cell Factories. 2015;14(1):11.
- Yi H, Cho YJ, Yong D, Chun J. Genome Sequence of Escherichia coli J53, a Reference Strain for Genetic Studies. Journal of Bacteriology. 2012;194(14):3742-3.
- Baumann RLB, E. H.; Wiseman, J. S.; Vaal, M.; Nichols, J. S. Inhibition of Escherichia coli Growth and Diaminopimelic Acid Epimerase by 3-Chlorodiaminopimelic Acid. Antimicrobial Agents and Chemotherapy 1988;32:1119-23.
- Rocha D, Santos, CS, Pacheco LG. Bacterial reference genes for gene expression studies by RT-qPCR: survey and analysis. Antonie Van Leeuwenhoek. 2015;108:685-93.
Transcript
Bacterial cells, such as E. coli, are able to transfer genetic information from cell-to-cell. Conjugation differs from other mechanisms of DNA transfer, such as transduction or transformation, in that it requires physical contact between the cells.
To proceed, conjugation requires a donor cell that expresses the fertility, or F, factor and a recipient cell without it, an F minus cell. The process requires two steps. The first is the establishment of direct cell-to-cell contact. To do this, the donor cell generates an extracellular filamentous structure called a sex pilus. It is named this since conjugation is a form of mating for asexually reproducing bacteria, but it should be noted that it is not true sexual reproduction as no gametes are exchanged and no offspring are formed.
The second step is delivery of DNA to the recipient cell. After the sex pilus establishes contact between two cells, a conduit called the Type IV secretion system is built allowing for the transfer of DNA. The donor cell then begins to replicate the extrachromosomal DNA that will be transferred selected based on the presence of a genetic element known as the OriT or origin of transfer. One end of the newly replicated DNA is threaded into the conduit through DNA protein binding. As the DNA is further replicated, it is pumped through the channel, facilitated by a complex of proteins encoded by genes located close to the OriT. Once the DNA is fully transferred, it will either form an extra chromosomal plasmid, or it may integrate into the chromosome of the recipient cell. Whichever the endpoint of the transferred DNA, the genes it encodes will then be expressed. This gene expression can be used to confirm successful conjugation.
For example, consider a scenario where the donor strain expresses ampicillin resistance and passes this on in the conjugated DNA to the recipient bacterium, but the recipient strain also has a tetracycline resistance gene not present in the donor. In this event, when the cells are plated on LB media containing both tetracycline and ampicillin, colonies should grow only from successfully conjugated bacteria, which will be expressing both resistance phenotypes. To further confirm successful conjugation, plasmid DNA from these colonies can be harvested and then a section of DNA specific to the transferred plasmid can be amplified using polymerase chain reaction, or PCR. When the PCR product is run on an electrophoresis gel alongside a ladder of standard sizes, a PCR fragment of a known size should be visible on the gel, further confirming successful conjugation. In this experiment, a plasmid will be used to transfer the ampicillin resistance gene via conjugation from a donor strain to a tetracycline-resistant recipient strain. After this, to confirm conjugation, the conjugation mixture will be incubated on a plate containing both antibiotics leaving only the transformed bacteria. Finally, successful conjugation will be further confirmed with PCR.
Before starting the procedure, put on the appropriate personal protective equipment, including a lab coat and gloves. Next, sterilize the workspace using 70% ethanol to wipe down the surface.
In this procedure, the ampicillin resistance gene will be transferred from the WM3064 strain of E. coli to the J53 strain of E. coli via conjugation. The donor strain WM3064 is resistant to tetracycline and ampicillin and it requires diaminopimelic acid, or DAP, to grow. The recipient strain J53 is only resistant to tetracycline and it does not require DAP to grow. This means that successfully conjugated cells should be resistant to tetracycline and ampicillin and can grow without DAP.
Prepare the donor strain culture by inoculating five milliliters of LB containing 0.3 millimoles of DAP with a scrap of the frozen donor strain glycerol stock. Then, prepare the recipient strain by inoculating five milliliters of LB broth without DAP with a scrap of the frozen recipient strain glycerol stock. Grow these cultures overnight at 37 degrees Celsius with aeration and shaking at 220 RPM in a shaking incubator. Once the cultures have grown to an OD 600 of two, remove one milliliter of culture from each and place this into two new separate 1.5 milliliter microcentrifuge tubes. Then, centrifuge these aliquots at 3000 RPM for five minutes to pellet the bacterial cells. Discard the supernatant and wash each pellet with 250 microliters of 1X PBS. Centrifuge the samples again and, after discarding the supernatant, resuspend each pellet in 500 microliters of PBS.
To begin the conjugation procedure, first combine 50 microliters of recipient cells with 50 microliters of donor cells in a 1.5 milliliter microcentrifuge tube and mix by pipetting up and down gently. Next, pipette 100 microliters of the recipient cell culture onto another 1X tetracycline plate containing DAP. Next, prepare your negative control by pipetting 100 microliters of the recipient cell culture only onto a non-selective agar plate containing DAP. Then, incubate the conjugation and negative control plates overnight at 37 degrees Celsius.
The next day, take a sterile cell scraper and harvest cells from the conjugation plate by collecting colonies. Then, transfer the colonies to a sterile 1.5 milliliter microcentrifuge tube containing one milliliter of 1X PBS. Repeat this process to collect the recipient cells from the other plate.
After this, vortex the samples to mix. After mixing, transfer the tubes to a centrifuge to gently pellet the cells. Discard the supernatant, then wash the cell pellets in one milliliter of PBS and vortex the tubes to resuspend the cells. Pellet the cells again by centrifuging. Discard the supernatant again and resuspend both cell pellets in one milliliter of PBS. Now, using a sterile pipette tip, plate 100 microliters of the conjugation reaction cell mixture onto an LB agar plate without DAP containing 1X tetracycline and 1X ampicillin. Repeat the plating method using 100 microliters of a ten-fold dilution of the same cell mixture in PBS onto another LB agar plate without DAP containing 1X tetracycline and 1X ampicillin.
Finally, pipette 100 microliters of the negative control cell mixture onto a single LB agar plate with 1X tetracycline only. After overnight incubation at 37 degrees Celsius, the colonies should be visible. Using a sterile pipette tip, pick a single colony from the conjugation reaction plate and add it to a tube containing five milliliters of selective LB media containing both antibiotics. Then, repeat the colony isolation by selecting a single colony from the recipient cell plate. Grow these cultures overnight at 37 degrees Celsius with aeration at 220 RPM.
The next day, wipe down the bench top with 70% ethanol and remove the plates from the incubator. Use a DNA mini prep kit to isolate DNA from 4. 5 milliliters of each culture according to the manufacturer’s instructions. After completing the DNA mini prep, elute the DNA using 35 microliters of nuclease-free water. Finally, use the remaining 0. 5 milliliters of each culture to prepare one milliliter glycerol stocks by adding 0.5 milliliters of 100% glycerol for a one-to-one dilution. Place these aliquots at minus 80 degrees Celsius for storage until needed.
To confirm successful conjugation by PCR, first prepare a PCR master mix by adding 75 microliters of 2X PCR master mix to a microcentrifuge tube. Then, add 7.5 microliters each of a 10 micromolar forward primer and a 10 micromolar reverse primer designed to amplify the ampicillin resistance gene from the plasmid. Next, prepare a second PCR master mix by adding 75 microliters of 2X PCR master mix to a microcentrifuge tube and then adding 7.5 microliters each of a 10 micromolar forward primer and 10 micromolar reverse primer designed to amplify a housekeeping gene, in this case DNA gyrase B.
Now, add 15 microliters of the first master mix to a PCR tube and then add 10 nanograms, approximately two microliters of the template experimental DNA to the same tube. Bring the reaction up to a final volume of 25 microliters with nuclease-free water. Repeat these steps to produce the remaining five reactions, so that the tubes contain the components shown here. Now, transfer these reactions to a thermocycler with the block pre-heated to 98 degrees Celsius and then initiate the program. After completion of the PCR, remove the tubes from the machine. Then, load two microliters of each reaction mixed with two microliters of loading dye and four microliters of a molecular weight marker into consecutive wells of a 1% agarose gel. Set the gel to run at 150 volts for 20 minutes. Finally, visualize the gel using a UV illuminator.
In this experiment, the successful transfer of the ampicillin resistance gene via conjugation was confirmed via PCR. Here, a roughly 500 base pair sized band should be observed in the well containing the conjugated DNA and ampicillin primers, well two in this example. A housekeeping gene, DNA gyrase B, was loaded into wells three and five with conjugated DNA and recipient cell DNA, respectively. Bands observed in these wells act as a positive control to ensure the DNA template was present and that PCR was successful. Bands should not be observed in the well containing the reaction for recipient cell DNA and the ampicillin primer pair, well four in this example, because the recipient cells are not ampicillin-resistant. Additionally, no bands should be observed in the reactions lacking template DNA, wells six and seven here. If these conditions are met, this will confirm the successful transfer of the ampicillin resistance gene, conferring ampicillin resistance from the WM3064 strain of E. coli to the J53 strain of E. coli.
