Rapid Isolation And Purification Of Mitochondria For Transplantation By Tissue Dissociation And Differential Filtration
Summary
A method for rapid isolation of mitochondria from mammalian tissue biopsies is described. Rat liver or skeletal muscle preparations were homogenized with a commercial tissue dissociator and mitochondria were isolated by differential filtration through nylon mesh filters. Mitochondrial isolation time is <30 min compared to 60 – 100 min using alternative methods.
Abstract
Previously described mitochondrial isolation methods using differential centrifugation and/or Ficoll gradient centrifugation require 60 to 100 min to complete. We describe a method for the rapid isolation of mitochondria from mammalian biopsies using a commercial tissue dissociator and differential filtration. In this protocol, manual homogenization is replaced with the tissue dissociator’s standardized homogenization cycle. This allows for uniform and consistent homogenization of tissue that is not easily achieved with manual homogenization. Following tissue dissociation, the homogenate is filtered through nylon mesh filters, which eliminate repetitive centrifugation steps. As a result, mitochondrial isolation can be performed in less than 30 min. This isolation protocol yields approximately 2 x 1010 viable and respiration competent mitochondria from 0.18 ± 0.04 g (wet weight) tissue sample.
Introduction
Mitochondria exist in every cell in the body except red blood cells and are involved in a large number of important cellular and metabolic processes1-4. Because of these many functions, mitochondrial damage can have detrimental effects3. In order to investigate mitochondrial function and dysfunction several mitochondrial isolation methods have been described. The earliest published accounts of mitochondrial isolation date to the 1940s5-8. The first documented attempt demonstrated mitochondrial isolation by grinding liver tissue in a mortar followed by centrifugation in a salt solution at low speed5,8. Later, other groups expanded upon the original procedure and demonstrated tissue fractionation based on differential centrifugation6-8. These early methods formed the basis of current techniques which often incorporate homogenization, and/or differential centrifugation9-15. The number of homogenization and centrifugation steps varies among protocols. These repetitive steps increase the time for mitochondrial isolation and ultimately reduce viability. In addition, manual homogenization can cause mitochondrial damage and inconsistent results if not properly controlled10,16.
Recently, we used homogenization and differential centrifugation to isolate mitochondria for transplantation into myocardial tissue17,18. This lengthy isolation procedure required approximately 90 min and the clinical applicability of this method was therefore limited. To allow for acute therapeutic use in clinical and surgical treatment we have developed a rapid mitochondrial isolation procedure that can be performed in less than 30 min.
The major benefits of this protocol are that standardized tissue dissociation allows for uniform and consistent homogenization of tissue that is not easily achieved with manual homogenization. In addition, the use of differential filtration in place of differential centrifugation eliminates time consuming and repetitive centrifugation steps allowing for more rapid isolation of highly purified, viable and respiration competent mitochondria.
The ability to isolate viable and respiration competent mitochondria in less than 30 min allows for clinical applicability. This isolation protocol has potential for use in coronary artery bypass grafting surgery (CABG) and other therapeutic procedures.
Protocol
Preparation
- Prepare 1 M K-HEPES Stock Solution (adjust pH to 7.2 with KOH).
- Prepare 0.5 M K-EGTA Stock Solution (adjust pH to 8.0 with KOH).
- Prepare 1 M KH2PO4 Stock Solution.
- Prepare 1 M MgCl2 Stock Solution.
- Prepare Homogenizing Buffer (pH 7.2) 300 mM sucrose, 10 mM K-HEPES, and 1 mM K-EGTA. Store at 4 °C.
- Prepare Respiration Buffer 250 mM sucrose, 2 mM KH2PO4, 10 mM MgCl2, 20 mM K-HEPES Buffer (pH 7.2) and 0.5 mM K-EGTA (pH 8.0). Store at 4 °C.
- Prepare 10x PBS Stock Solution by dissolving 80 g of NaCl, 2 g of KCl, 14.4 g of Na2HPO4, and 2.4 g of KH2PO4 into 1 L double distilled H2O (pH 7.4).
- Prepare 1x PBS by pipetting 100 ml 10x PBS into 1 L double distilled H2O.
- Prepare Subtilisin A Stock by weighing out 4 mg of Subtilisin A into a 1.5 ml microfuge tube. Store at -20 °C until use.
- Prepare BSA Stock by weighing out 20 mg of BSA into a 1.5 ml microfuge tube. Store at -20 °C until use.
Mitochondrial Isolation (Figure 1)
- Immediately prior to isolation, dissolve Subtilisin A in 1 ml of Homogenizing Buffer.
- Immediately prior to isolation, dissolve BSA in 1 ml of Homogenizing Buffer.
- Collect two fresh tissue samples using a 6 mm biopsy sample punch and store in 1x PBS in a 50 ml conical centrifuge tube on ice.
- Transfer the two 6 mm punches of tissue to a dissociation C tube containing 5 ml of ice cold Homogenizing Buffer.
- Homogenize the tissue by fitting the dissociation C tube on the tissue dissociator and select the pre-set mitochondrial isolation cycle (60 sec homogenization).
- Remove the dissociation C tube to an ice bucket.
- Add 250 μl of Subtilisin A Stock Solution to the homogenate, mix by inversion and incubate the homogenate on ice for 10 min.
- Optional in the case that the tissue is fibrous: Centrifuge the solution at 750 x g for 4 min.
- Place a 40 µm mesh filter onto a 50 ml conical centrifuge tube on ice and pre-wet the filter with Homogenizing Buffer and filter the homogenate into the 50 ml conical centrifuge tube on ice.
- Add 250 µl of freshly prepared BSA Stock Solution to the filtrate and mix by inversion.
Note: Omit this step if mitochondrial protein determination is required. - Place a 40 µm filter onto a 50 ml conical centrifuge tube on ice and pre-wet the filter with Homogenizing Buffer and filter the homogenate into the 50 ml conical centrifuge tube on ice.
- Place a 10 µm filter onto a 50 ml conical centrifuge tube on ice and pre-wet the filter with Homogenizing Buffer and filter the homogenate into the 50 ml conical centrifuge tube on ice.
- Transfer the filtrate to two pre-chilled 1.5 ml microfuge tubes and centrifuge at 9,000 x g for 10 min at 4 ºC.
- Remove supernatant and re-suspend and combine pellets in 1 ml of ice cold Respiration Buffer.
ATP Assay
Note: To determine the metabolic activity of isolated mitochondria an ATP luminescence assay can be performed using an ATP assay kit. The protocol, reagents and standards were supplied in the assay kit. A summary of the procedure is described below.
- Equilibrate kit reagents to RT.
- Prepare 10 mM ATP Stock Solution by dissolving lyophilized ATP pellet in 1,170 µl of double distilled water. Store ATP standard Stock Solution and prepared mitochondrial samples on ice.
- Add 5 ml of Substrate Buffer solution to a vial of lyophilized substrate solution. Mix gently and place in the dark.
- Add 100 µl of Respiration Buffer to all wells of a black, opaque bottom, 96 well plate.
- Add 10 µl of mitochondria from the prepared samples to each well of a black, opaque bottom, 96 well plate. Note: Samples are plated in triplicate. Include a row for standards and three wells for the negative control (Respiration Buffer).
- Add 50 µl of mammalian cell lysis solution to all wells, including standards and controls.
- Incubate the plate at 37 ºC for 5 min on an orbital shaker at 125 rpm.
- During the incubation prepare ATP standards in concentrations of 0.1 mM, 0.05 mM, 0.01 mM, 0.005 mM, 0.001 mM, and 0.0001 mM ATP from the 10 mM ATP Stock Solution. Store standards on ice.
- Following the incubation, add 10 µl of ATP standards to corresponding wells as indicated on the plate map. Note: Standards are performed in duplicate.
- Add 50 µl of the reconstituted substrate solution to each well.
- Incubate the plate at 37 ºC on the orbital shaker for 5 min at 125 rpm.
- Open Gen5 1.11 software on a computer linked with the spectrophotometer.
- Under “Create a New Item”, click on “Experiment”.
- Click on “Default Protocol”. Click “Ok”.
- In the left column, select “Protocol”, then “Procedure”.
- Select “Delay”. Set to 00:10:00. Click “Ok”.
- Select “Read”. Select “Luminescence” from drop down menu. Adjust other settings to the following: Read type Endpoint, Integration Time 0:01.00 MM:SS.ss, Filter Sets 1, Emission Hole, Optics Position Top, Sensitivity 100, Top Probe Vertical Offset 1.00 mm.
- When the plate is ready to be analyzed, click the “Read Plate” icon on the top row. Click “Read”. When the spectrophotometer opens, place the plate into the tray with well A1 in the upper left corner. A box with the temperature will open. Click “Read”. Note: Higher values correlate with increased ATP levels and higher metabolic activity.
Representative Results
A figure outlining the procedural steps in the isolation of mitochondria using tissue dissociation and differential filtration is shown in Figure 1. Total procedural time is less than 30 min.
Tissue samples were obtained using a 6 mm biopsy punch. Tissue weight was 0.18 ± 0.04 g (wet weight). The number of mitochondria isolated as determined by particle size counting was 2.4 x 1010 ± 0.1 x 1010 mitochondria for skeletal muscle and 2.75 x 1010 ± 0.1 x 1010 mitochondria for liver preparations (Figure 2A). To allow for comparison mitochondrial number was also determined by hemocytometer. Mitochondrial number was underestimated as determined by hemocytometer as 0.11 x 1010 ± 0.04 x 1010 mitochondria for skeletal muscle and 0.34 x 1010 ± 0.09 x 1010 mitochondria for liver preparations (Figure 2A). Mitochondrial diameter as determined by size based particle counter is shown in Figure 2B. The representative tracing shows the isolated mitochondria are localized under one peak with mean diameter of 0.38 ± 0.17 µm in agreement with previous reports7.
Mitochondrial protein/g (wet weight) starting tissue as determined by Bicinchoninic Acid (BCA) assay was 4.8 ± 2.9 mg/g (wet weight) and 7.3 ± 3.5 mg/g (wet weight) for skeletal muscle and liver samples respectively (Figure 2C).
Mitochondrial purity was determined by transmission electron microscopy and is shown in Figure 2D. Mitochondria are shown to be electron dense with less than 0.01% being fractured or damaged. Contamination by non-mitochondrial particles is less than 0.001%.
Mitochondrial viability was determined by MitoTracker Red as previously described17,18. Our results show that the isolated mitochondria maintain membrane potential (Figures 3A – C).
ATP was determined using a luminescent assay kit. A plate map for the ATP assay is shown in Figure 4. ATP standards were plated in duplicate. Mitochondrial samples and negative controls were plated in triplicate. ATP content was 10.67± 4.38 nmol/mg mitochondrial protein and 14.83 ± 4.36 nmol/mg mitochondrial protein for skeletal muscle and liver samples respectively (Figure 3D).
Mitochondrial respiration was assessed using a Clark type electrode as previously described17,18. Mitochondrial oxygen consumption rate was 178 ± 17 nM O2/min/mg mitochondrial protein for skeletal muscle and 176 ± 23 nM O2/min/mg mitochondrial protein for liver preparations. Respiratory control index (RCI) values were 2.45 ± 0.34 and 2.67 ± 0.17 for skeletal muscle and liver sample preparations respectively (Figure 3E). These results are similar to those reported in our previous studies using manual homogenization and differential centrifugation to isolate mitochondria17-18.
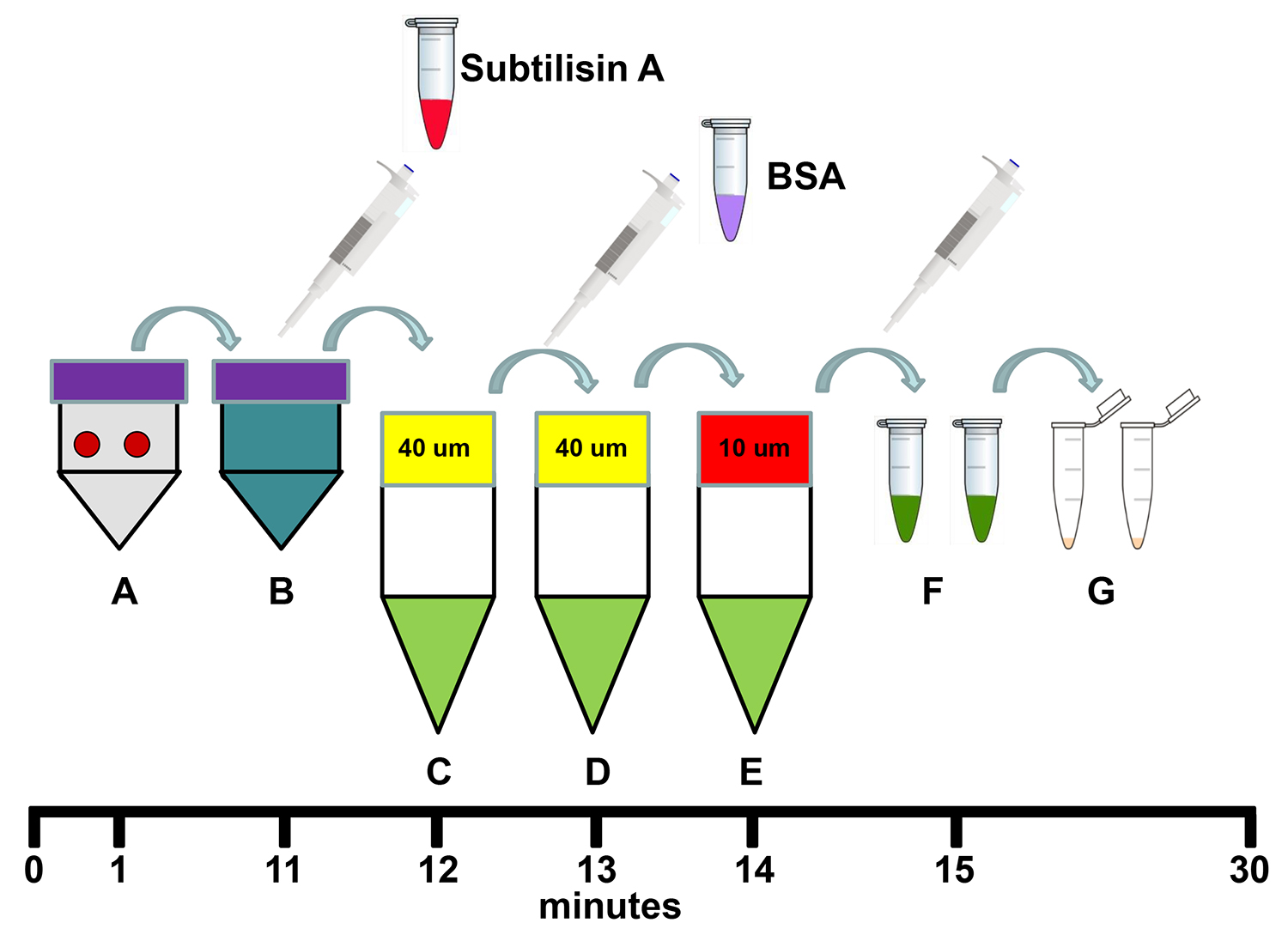
Figure 1. Schema for the isolation of mitochondria using tissue dissociation and differential filtration. (A) Transfer two 6 mm biopsy sample punches to 5 ml of Homogenizing Buffer in a dissociation C tube and homogenize the samples using the tissue dissociator's 1 min homogenization program. (B) Add 250 µl Subtilisin A stock solution to the homogenate in the dissociation C tube and incubate on ice for 10 min. (C) Filter the homogenate through a pre-wetted 40 µm mesh filter in a 50 ml conical centrifuge tube on ice and then add 250 µl of BSA stock solution to the filtrate. (D) Re-filter the filtrate through a new pre-wetted 40 µm mesh filter in a 50 ml conical centrifuge on ice. (E) Re-filter the filtrate through a new pre-wetted 10 µm mesh filter in a 50 ml conical centrifuge tube on ice. (F) Transfer the filtrate to 1.5 ml microfuge tubes and centrifuge at 9,000 x g for 10 min at 4 °C. (G) Remove the supernatant and re-suspend and combine the mitochondrial pellets in 1 ml Respiration Buffer. Total procedure time is less than 30 min. Please click here to view a larger version of this figure.
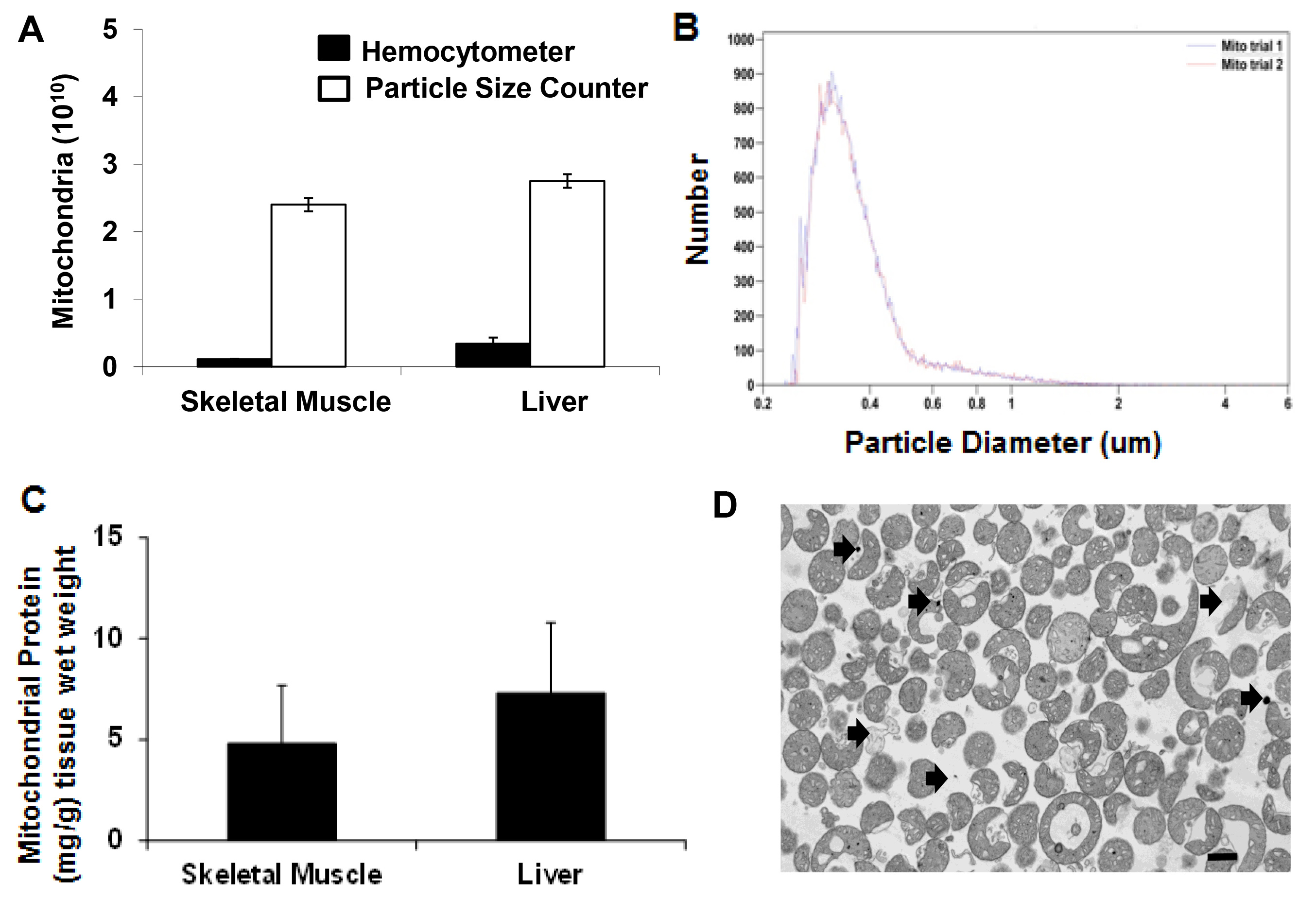
Figure 2. Mitochondrial yield and purity. (A) Hemocytometer and particle size counter mitochondria number isolated from 0.18 ± 0.04 g tissue (wet weight) for skeletal muscle and liver. (B) Mitochondrial size (mg/g) distribution as detected by particle size counter. (C) Mitochondrial protein mg/g tissue wet weight for skeletal muscle and liver. (D) Transmission electron microscopy image of isolated mitochondria. Scale bar is 100 nm. Arrows indicate possible contamination by non mitochondrial particles and damaged mitochondria. Please click here to view a larger version of this figure.
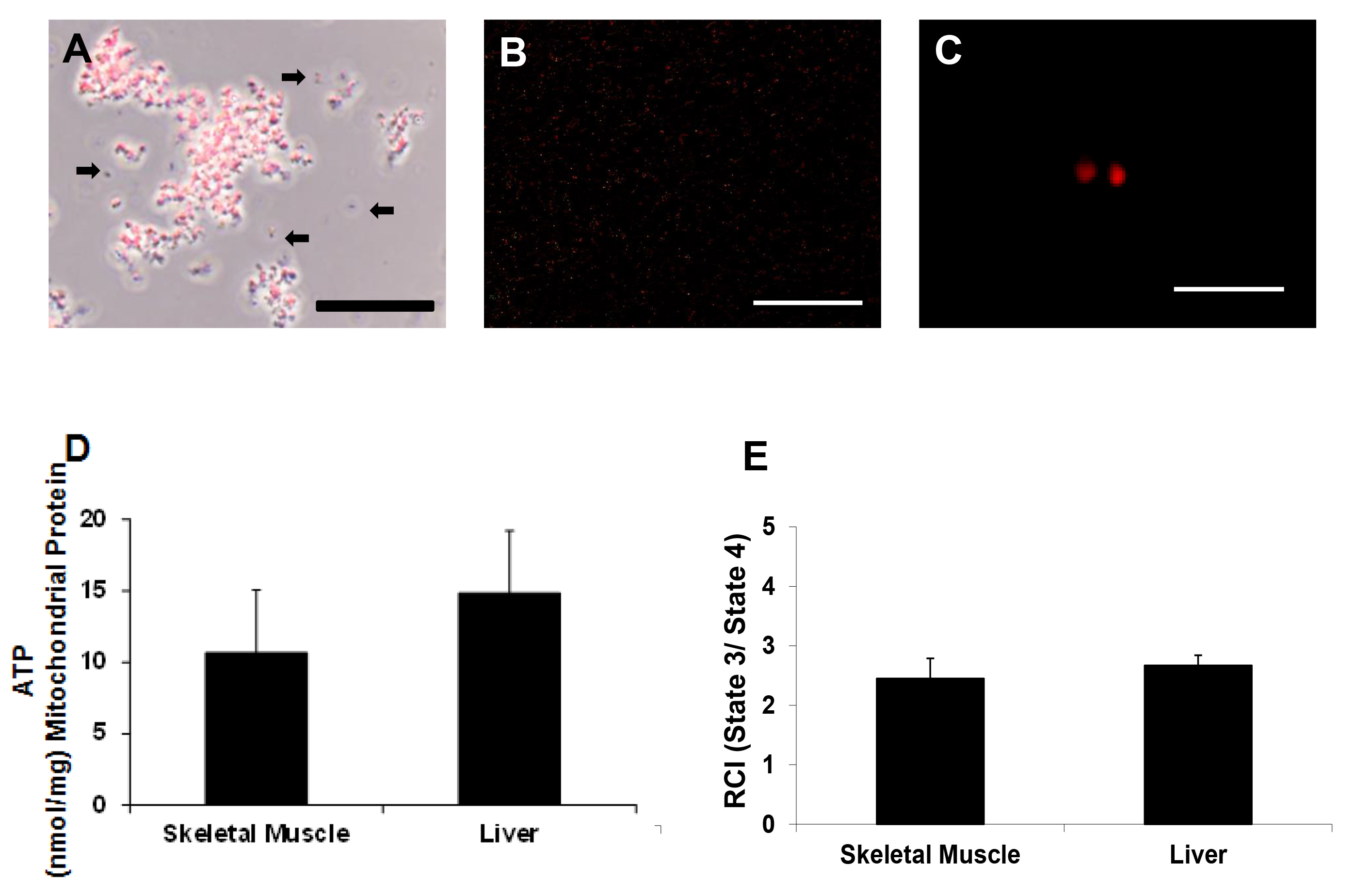
Figure 3. Mitochondrial viability. Representative photomicrographs of isolated mitochondria (A) under phase contrast illumination and (B and C) under fluorescence, with mitochondria labeled with MitoTracker Red CMXRos. Scale bars are 25 μm (A, B) and 5 μm (C). These images indicate that mitochondria maintained membrane potential. Arrows indicate mitochondria lacking membrane potential or debris (D) ATP content nmol/mg mitochondrial protein as determined by ATP assay and (E) RCI (state 3/state 4) as determined by Clark electrode. Please click here to view a larger version of this figure.

Figure 4. Plate map for ATP assay. This plate map illustrates how to set up standards (A1 – A12), mitochondria samples (B1 – C6), and negative controls (C7 – C9) for the ATP assay. During the assay, 100 µl of Respiration Buffer, 50 µl of mammalian cell lysis solution and 50 µl of reconstituted substrate solution are added to all wells (A1 – C9).
Discussion
To successfully isolate mitochondria using this protocol it is essential to keep all solutions and tissue samples on ice to preserve mitochondrial viability. Even when maintained on ice, isolated mitochondria will exhibit a decrease in functional activity over time19. We recommend that all solutions and additions be pre-prepared. We pre-weigh and store Subtilisin A in 4 mg aliquots in 1.5 ml microfuge tubes and store them at -20 °C. Similarly BSA is pre-weighed and stored in 20 mg aliquots in 1.5 ml microfuge tubes that can be stored at -20 °C. Just prior to use the tubes are removed from -20 °C and Subtilisin A and BSA are dissolved in 1 ml of Homogenizing Buffer for use in the mitochondrial isolation procedure.
Two biopsy punches from skeletal muscle tissue obtained using a 6 mm biopsy punch provide sufficient mitochondria for use in clinical and surgical procedures for therapeutic interventions17-18. To estimate the number of mitochondria we have used two methods, hemocytometer and particle size counting. The use of particle size counting is recommended. The particle size counter uses electrical impedance to measure the volume of particles passing through an aperture of a defined size. The particle size counter is costly but provides accurate and reliable estimates and is user independent. Mitochondrial number estimated by hemocytometry is more economical. Our studies have demonstrated that this method provides variable estimates that are approximately one order of magnitude less than that obtained using a particle size counter. We have also noted that counts obtained by hemocytometer are highly dependent on the user. We suggest that to ensure consistent estimates all counts using a hemocytometer should be performed by one person.
We have found ATP assay kits to be useful for determining mitochondrial function. The kit supplies all necessary reagents and provides a simple and fast method for determining the metabolic activity of isolated mitochondria. The ATP assay provides similar results as those obtained using a Clark-type electrode and therefore is compatible with previous data analysis20-21.
A major advantage of our mitochondrial isolation protocol is that it allows for isolation of a high yield of viable, respiration competent mitochondria free of contamination in less than 30 min. Differential filtration in place of differential centrifugation significantly reduces procedure time. Other protocols incorporate several centrifugation steps with overall isolation time being 60 min to 100 min13-17. Another advantage of this protocol is that tissue homogenization is standardized. The commercial tissue dissociator provides a standardized cycle and yields consistent and reproducible results. This is in contrast to manual homogenization that is subject to user variability and inconsistency. Our method for the rapid isolation of mitochondria using tissue dissociation and differential filtration provides an isolation time frame compatible for clinical and surgical therapeutic intervention17,18.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This study was supported by National Heart, Lung, and Blood Institute Grant HL- 103542, and The BCH Anesthesia Research Foundation’s Distinguished Trailblazer Award to CAP.
Materials
| Sucrose | Sigma Aldrich | 84100 | |
| HEPES | Sigma Aldrich | H4034 | |
| EGTA | Sigma Aldrich | E4378 | |
| Substilsin A | Sigma Aldrich | P5380 | |
| BSA | Sigma Aldrich | A7906 | |
| KH2PO4 | Sigma Aldrich | P5379 | |
| MgCl2 | Sigma Aldrich | M8266 | |
| NaCl | Sigma Aldrich | S6191 | |
| KCl | Fisher Scientific | P2173 | |
| Na2HPO4 | Fisher Scientific | S374 | |
| ATPlite Luminescence Assay System, 1000 Assay Kit |
Perkin Elmer | 6016941 | |
| Equipment | |||
| 50 mL Conical Tubes | BD | 352098 | |
| 40 μm Nylon Filters | BD | 352340 | |
| GentleMACS C tube | Miltenyl Biotech | 120-005-331 | |
| 1.5 mL Eppendorf tube | Fisher Scientific | 05-408-129 | |
| 6 mm biopsy punch | Miltex | 33-36 | |
| 10 μm Pluristrainer | Pluriselect | 43-500-10-03 | |
| Eppendorf Centrifuge 5415C | Marshall Scientific | EP-5415C | |
| GentleMACS Dissociator | Miltenyl Biotech | 130-093-235 | |
| 96-well plates, tissue culture treated | VWR | 82050-732 | |
| Rotomax 120 orbital shaker | Heidolph | 544-41200-00 | |
| Synergy H4 Hybrid Multi-Mode Microplate Reader |
BioTek | ||
| Multisizer 4 Coulter Counter | Beckman Coulter | A63076 | |
| Oxytherm System | Hansatech Instruments | ||
| Hemacytometer | Fisher Scientific | 267110 |
References
- van Loo, G., Saelens, X., van Gurp, M., MacFarlane, M., Martin, S. J., Vandenabeele, P. The role of mitochondrial factors in apoptosis: a Russian roulette with more than one bullet. Cell Death Differ. 9 (10), 1031-1042 (2002).
- Szabadkai, G., Duchen, M. R. Mitochondria: the hub of cellular Ca2+ signaling. Physiology (Bethesda). 23, 84-94 (2008).
- Chan, D. C. Mitochondria: Dynamic Organelles in Disease, Aging, and Development. Cell. 125 (7), 1241-1252 (2006).
- Picard, M., et al. Mitochondrial structure and function are disrupted by standard Isolation methods. PLoS ONE. 6 (3), e18317 (2011).
- Bensley, R. R., Hoerr, N. Studies on cell structure by freeze-drying method; preparation and properties of mitochondria. Anat Rec. 60, 449-455 (1934).
- Claude, A. Fractionation of mammalian liver cells by differential centrifugation II. Experimental procedures and results. J Exp Med. 84 (1), 61-89 (1946).
- Hogeboom, H., Schneider, W. C., Palade, G. E. Cytochemical studies of mammalian tissues; isolation of intact mitochondria from rat liver; some biochemical properties of mitochondria and submicroscopic particulate material. J Biol Chem. 172 (2), 619-635 (1948).
- Ernster, L., Schtaz, G. Mitochondria: a historical Review. J Cell Biol. 91 (3 Pt 2), 227s-255s (1981).
- Pallotti, F., Lenaz, G. Isolation and subfractionation of mitochondria from animal cells and tissue culture lines. Methods Cell Biol. 80, 3-44 (2007).
- Schmitt, S., et al. A semi-automated method for isolating functionally intact mitochondria from cultured cells and tissue biopsies. Anal Biochem. 443 (1), 66-74 (2013).
- Fernández-Vizarra, E., Ferrín, G., Pérez-Martos, A., Fernández-Silva, P., Zeviani, M., Enríquez, J. A. Isolation of mitochondria for biogenetical studies: An update. Mitochondrion. 10 (3), 253-262 (2010).
- Graham, J. M. Chapter 3, Unit 3.3, Isolation of mitochondria from tissues and cells by differential centrifugation. Curr Protoc Cell Biol. , (2001).
- Frezza, C., Cipolat, S., Scorrano, L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc. 2 (2), 287-295 (2007).
- Wieckowski, M. R., Giorgi, C., Lebiedzinska, M., Duszynski, J., Pinton, P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat Protoc. 4 (11), 1582-1590 (2009).
- Gostimskaya, I., Galkin, A. Preparation of highly coupled rat heart mitochondria. J Vis Exp. (43), (2010).
- Gross, V. S., et al. Isolation of functional mitochondria from rat kidney and skeletal muscle without manual homogenization. Anal Biochem. 418 (2), 213-223 (2011).
- Masuzawa, A., et al. Transplantation of autologously derived mitochondria protects the heart from ischemia-reperfusion injury. American J Physiol Heart and Circ Physiol. 304 (7), (2013).
- McCully, J. D., et al. Injection of isolated mitochondria during early reperfusion for cardioprotection. American J Physiol Heart and Circ Physiol. 296 (1), 94-105 (2009).
- Olson, M. S., Von Korff, R. W. Changes in endogenous substrates of isolated rabbit heart mitochondria during storage. J Biol Chem. 242 (2), 325-332 (1967).
- Diepart, C., et al. Comparison of methods for measuring oxygen consumption in tumor cells in vitro. Anal Biochem. 396 (2), 250-256 (2010).
- Lanza, I. R., Nair, K. S. Functional assessment of isolated mitochondria in vitro. Methods Enzymol. 457, 349-372 (2009).

