A Strategy for Sensitive, Large Scale Quantitative Metabolomics
Summary
Metabolite profiling has been a valuable asset in the study of metabolism in health and disease. Utilizing normal-phased liquid chromatography coupled to high-resolution mass spectrometry with polarity switching and a rapid duty cycle, we describe a protocol to analyze the polar metabolic composition of biological material with high sensitivity, accuracy, and resolution.
Abstract
Metabolite profiling has been a valuable asset in the study of metabolism in health and disease. However, current platforms have different limiting factors, such as labor intensive sample preparations, low detection limits, slow scan speeds, intensive method optimization for each metabolite, and the inability to measure both positively and negatively charged ions in single experiments. Therefore, a novel metabolomics protocol could advance metabolomics studies. Amide-based hydrophilic chromatography enables polar metabolite analysis without any chemical derivatization. High resolution MS using the Q-Exactive (QE-MS) has improved ion optics, increased scan speeds (256 msec at resolution 70,000), and has the capability of carrying out positive/negative switching. Using a cold methanol extraction strategy, and coupling an amide column with QE-MS enables robust detection of 168 targeted polar metabolites and thousands of additional features simultaneously. Data processing is carried out with commercially available software in a highly efficient way, and unknown features extracted from the mass spectra can be queried in databases.
Introduction
Metabolomics, defined as an experiment that measures multiple metabolites simultaneously, has been an area of intense interest. Metabolomics provides a direct readout of molecular physiology and has provided insights into development and disease such as cancer1-4. Nuclear magnetic resonance (NMR) and gas chromatography-mass spectrometry (GC-MS) are among the most commonly used instruments5-9. NMR, especially has been used for flux experiments since heavy isotope labeled compounds, such as 13C labeled metabolites, are NMR-active10,11. However, this strategy requires relatively high sample purity and large sample quantity, which limits its applications in metabolomics. Meanwhile, data collected from NMR needs intensive analysis and compound assignment of complex NMR spectra is difficult. GC-MS has been widely used for polar metabolites and lipid studies, but it requires volatile compounds and therefore often derivatization of metabolites, which sometimes involves complex chemistry that can be time consuming and introduces experimental noise.
Liquid chromatography (LC) coupled to triple quadrupole mass spectrometry uses the first quadrupole for selecting the intact parent ions, which are then fragmented in the second quadrupole, while the third quadrupole is used to select characteristic fragments or daughter ions. This method, which records the transition from parent ions to specific daughter ions, is termed multiple reaction monitoring (MRM). MRM is a very sensitive, specific, and robust method for both small molecule and protein quantitation12-15,21. However, MRM does have its limitations. To achieve high specificity a MRM method needs to be built for each metabolite. This method consists of identifying a specific fragment and corresponding optimized collision energy, which requires pre-knowledge of the properties of the metabolites of interest, such as chemical structure information. Therefore, with some exceptions involving the neutral loss of common fragments, it is not possible to identify unknown metabolites with this method.
In the recent years, high-resolution mass spectrometry (HRMS) instruments have been released, such as the LTQ-orbitrap and Exactive series, the QuanTof, and TripleTOF 560016-18,22. HRMS can provide a mass to charge ratio (m/z) of intact ions within an error of a few ppm. Therefore, an HRMS instrument operated by detecting all precursor ions (i.e. full scan mode) can obtain direct structural information from the exact mass and the resulting elemental composition of the analyte, and this information can be used to identify potential metabolites. Indeed, all information about a compound can be obtained with an exact mass, up to the level of structural isomers. Also, a full scan method does not require previous knowledge of metabolites and does not require method optimization. Moreover, since all ions with m/z falling into the scan range can be analyzed, HRMS has a nearly unlimited capacity in terms of the number of metabolites that can be quantified in a single run compared to the MRM method. HRMS is also comparable to a triple quadrupole MRM in quantitative capacity due to the short duty cycle resulting in a comparable number of data points that can be obtained in a full MS scan. Therefore, HRMS provides an alternative approach for quantitative metabolomics. Recently, an improved version of HRMS termed Q-Exactive mass spectrometry (QE-MS) can be operated under the switching between positive and negative modes with sufficiently fast cycle times in a single method, which expands the detection range19. Here we describe our metabolomics strategy using the QE-MS.
Protocol
1. Preparation of LC-MS Reagents, Establishment of a Chromatography Method, and Establishment of Instrument Operating Procedures
- Preparation of LC Solvents
- Prepare 500 ml mobile phases. A is 20 mM ammonium acetate and 15 mM ammonium hydroxide in 3% acetonitrile/water, final pH 9.0; and B is 100% acetonitrile.
- Loosely cap the bottle, place it in a water bath sonicator, and sonicate for 10 min without extra heating. (This step is to ensure that all of the ammonium salts completely dissolve and that there are no residual air bubbles.)
- Transfer 250 ml of the solvent to a 250 ml glass bottle for LC-MS use, and keep the remainder at 4 °C.
- Prepare the low mass range calibration solution. It is important to use a customized low mass range calibration mixture for metabolomics applications to ensure that accurate masses are detected at low molecular weights.
- Weigh 5 mg of both sodium fluoroacetate and homovanillic acid and dissolve them into 5 ml water to make a final concentration of 1 mg/ml. Dissolve diazinon in methanol to make a final concentration of 10 mg/ml.
- To prepare 1 ml of negative low mass calibration solution, mix 960 ml of thermo negative calibration solution with 20 ml of sodium fluoroaceate and homovanillic acid solution. To make 1 ml of positive low mass calibration solution, mix 990 ml of thermo positive calibration solution and 10 ml diazinon solution. (The low mass calibration solution should be stored at 4 °C and be prepared fresh every 2 months.)
- Calibration of QE-MS at a Low Mass Range
- Before performing low mass range calibration, carry out a standard mass calibration (m/z, 150-2,000) in both positive and negative modes based on the manufacturer’s instructions.
- Once a regular mass calibration passes, adjust the scan range to 60-900 m/z in the instrument control panel and a source CID of 25 eV for positive mode and 35 eV for negative mode is applied. (This will give robust signals of the caffeine fragment ion and the sulfate ion. The scan range here is fixed, because the last m/z shouldn’t be larger than 15x of the starting m/z)
- Once the ion source is stable, then perform the customized calibration. Note: A stable source is defined as less than 10% of the total ion current variation in positive mode, and less than 15% in negative mode. The customized calibration ions and corresponding m/z are listed in Table 1.
- Establish the LC-MS instrumentation for polar metabolite analysis. LC is coupled to a QE-MS for metabolite separation and detection.
- Equip the QE-MS with a Heated electrospray ionization probe (H-ESI). Set the relevant tuning parameters for the probe as listed: heater temperature, 120 °C; sheath gas, 30; auxiliary gas, 10; sweep gas, 3; spray voltage, 3.6 kV for positive mode and 2.5 kV for negative mode. Set the capillary temperature at 320 °C, and S-lens at 55.
- Build a full scan method as follows: Full scan range: 60 to 900 (m/z); resolution: 70,000; maximum injection time: 200 msec with typical injection times around 50 msec; automatic gain control (AGC): 3,000,000 ions. These settings result in a duty cycle of around 550 msec to carry out scans in both positive and negative mode.
- Establish the chromatography method. Employ an amide column (100 x 2.1 mm i.d., 3.5 mm) for compound separation at room temperature13,15. The mobile phase A is as described above, and mobile phase B is acetonitrile. Use a linear gradient as follows: 0 min, 85% B; 1.5 min, 85% B, 5.5 min, 35% B; 10min, 35% B, 10.5 min, 35% B, 14.5 min, 35% B, 15 min, 85% B, and 20 min, 85% B. The flow rate is 0.15 ml/min from 0 to 10 min and 15 to 20 min, and 0.3 ml/min from 10.5 to 14.5 min.
2. Preparation of Metabolite Samples
- Prepare an extraction solvent. Mix 40 ml methanol (LC-MS grade) and 10 ml water (LC-MS grade) in a 50 ml tube, and keep it in -80 °C freezer for at least 1 hr before use. Note: This procedure and the steps below can be modified for the extraction of biological tissue and fluid samples.
- Culture colon cancer HCT 8 cells in three 10 cm dishes or 6-well plates with full growth medium, RPMI 1640 supplemented with 10% heat inactivated Fetal Bovine Serum and 100,000 units/L penicillin and 100 mg/L streptomycin.
- When cells reach 80% confluence, quickly remove the medium, and place the dish or plate on top of dry ice13,15. Add 1 ml extraction solvent immediately (80% methanol/water), and transfer the plate to the -80 °C freezer. For a 10 cm dish, add 3 ml of extraction solvent to each well. (Try to remove the medium as much as possible to avoid the ion suppression effect due to residual salts from medium.)
- Leave the plate for 15 min. Remove it from the freezer, and scrape cells into the solvent on dry ice. Transfer the solution to 1.7 ml Eppendorf tubes, and centrifuge with the speed of 20,000 x g at 4 °C for 10 min. (Prepare cell metabolites from three separate dishes to make three replicate samples. The purpose of keeping two tubes is to have one as a backup.)
- Transfer the supernatant to two new Eppendorf tubes, and dry them in a speed vacuum. This takes about 3-6 hr depending on the speed vacuum used. (The samples can also be dried overnight under nitrogen gas.)
- After drying, store tubes of each sample in the -80 °C freezer. When ready, reconstitute one sample into 20 ml water (LC-MS grade), and inject 5 ml to LC-QE-MS for analysis.
3. Setup of Sample Sequence
- Once the calibration has been properly carried out on the QE-MS, equilibrate LC column for 5 min with 85% A at a flow rate of 0.15 ml/min, which is the starting condition of the LC gradient.
- Set up the sample sequence in random order. Note: In this way, it distributes the fluctuations introduced by the LC-MS to each sample and ensures more accurate comparison between different samples. Every 6 samples, add a wash run, which shares the same MS method, except the LC gradient is 95% A for 10 min and followed by a 5 min column equilibration at 85% A with a flow rate of 0.15 ml/min. Add blank samples (100% water) after each wash run to assess the system background and carry over levels.
- Save the sequence and once the LC column shows stable pressure, around 400 psi, start the sequence run. If there is no other sample is to be run after this sequence, then add a stop run in the end of the sequence, which has a flow rate of 0 ml/min in the end of the gradient and choose “standby” after finishing the sequence.
- Re-run the same sample set 12 hr after calibration. (This is to assess the mass error fluctuation after calibration.)
4. Post Analysis Instrument Cleaning and Maintenance
- At the end of sequence, wash the column with 95% A at a flow rate of 0.2 ml/min for 2 hr, and if necessary, reverse the column before washing.
- Remove the LC column and directly connect the LC to the ion source by a union. Prepare cleaning solvent, water/methanol/formic acid (v:v:v, 90:10:0.2), set MS on standby mode, and wash LC-MS system at a flow rate of 0.1 ml/min for 1 hr to remove the residual precipitated salts or other impurities. Lower the flow rate if there is too much system pressure.
- Set the capillary temperature at 50 °C, and remove the ion cage. Carefully take out the ion sweep cone and the ion transfer tube after the capillary temperature drops to 50 °C. Use a rough mesh, such as sandpaper, to remove impurities left on the surface of the ion sweep cone.
- Place the ion transfer tube into a 15 ml Falcon tube containing 10 ml 90% water/methanol with 0.1% formic acid. After sonicating the tube in a water bath sonicator for 20 min, decant the solvent inside, replace it with 10 ml pure methanol and sonicate for another 20 min. (If necessary, the sonication can be done at 40 °C or an even higher temperature to achieve better cleaning results.)
5. Analysis of LC-MS Data
- To ensure the sample sequence runs smoothly, after finishing the first two samples in the sequence, check peaks for unknown metabolites. Use a csv file listing metabolite names, neutral chemical formula and detection mode (either positive or negative), as the input file, and the output file contains extracted peaks and mass error in ppm. If the peak shape is abnormal or the mass error is off by more than 5 ppm, then the rest of the sequence needs to be stopped and troubleshooting needs to be done.
- Choose the method of “peak alignment and frame extraction” on commercially available software. Select raw data from LC-MS and group them. Pick samples in the middle of the run sequence as chromatography reference sample for peak alignment. Upload a frame seed including known metabolites for the targeted metabolites analysis with data collected and the corresponding frame seed.
- Perform data analysis in positive and negative mode separately. Use the default setting for other parameters. Turn off the database search function and run the workflow. Export the processed data as an excel sheet containing peak area of every frame. The first sets of frames correspond to the metabolites in the targeted list. Note: For a targeted metabolite analysis, the metabolites information is obtained based on the previous studies13,15.
- For an untargeted metabolite analysis, choose the method of “component extraction”. Load blank samples for background subtraction. Set peak intensity threshold at 105, m/z width of 10 ppm and signal to noise ratio of 3.
- Use human metabolome database for unknown compounds identification. Use a CV filter to remove the components with large CVs within replicate samples. Manually go through each component and pick those with well-defined peak or relatively big difference in different samples types for database search. Export data with hits in database. (Peak alignment could be bypassed if the peak alignment score is too low.)
Representative Results
The accuracy of metabolomics data highly depends on the LC-QE-MS instrument performance. To assess whether the instrument is operating in good condition, and whether the method applied is proper, several known metabolite LC peaks are extracted from the total ion chromatography (TIC), as shown in Figure 1. Polar metabolites, including amino acids, glycolysis intermediates, TCA intermediates, nucleotides, vitamins, ATP, NADP+ and so on have good retention on the column and good peak shapes in the amide column under current LC conditions. Meanwhile, a mass error test is done within 24 hr after low mass calibration, as illustrated in Figure 2. 6 different concentrations of samples in triplicate are run twice after calibration, and the whole time range covers almost 24 hr. The mass error is assessed by comparing the detected m/z to the theoretical m/z of targeted metabolites. Here the targeted metabolites have an m/z ranging from 74 (glycine) to 744 (NADP+). The Y axis here represents the accumulative percentage of metabolites within certain mass error range. The blue curve shows the result from 0-12 hr, while the red colored curve shows the data collected from 12-24 hr. Figure 2 clearly indicates that more than 90% of metabolites are within 5 ppm mass error, which means the low mass range calibration method developed here is sufficient to maintain 5 ppm mass error for low mass range detection.
Another issue to be addressed is the sensitivity of the instrument with the current method and instrument setup. A serial dilution of triplicate samples from 10 cm Petri dish was done 5 times with a dilution factor of 6, ending up with 6 different concentrations of samples. These samples represent the amount of metabolites extracted from 107, 1.67 x 106, 2.78 x 105, 4.63 x 104, 7.72 x 103, and 1.29 x 103 of cells, respectively. Since each concentration of sample is prepared in triplicate, a total of 18 samples are analyzed in LC-QE-MS. A targeted list is used to assess the number of metabolites detected at for differing concentration of sample. The result in Figure 3 indicates that the optimal number of targeted metabolites detected is between 2.78 x 105 and 1.67 x 106 cells, while 1 x 107 cells give a fewer number of detected metabolites, which is due to ion suppression effects. This result indicates that the optimal amount of cells to extract for this analysis is roughly that of a well of in a 6-well plate.
For untargeted metabolite analysis, a CV cutoff of 20% and an average intensity value of 107 are used to filter the components table. These rigid CV and average intensity threshold values are used for this demonstration aim. To improve reproducibility, CV cutoff values can be increased (for example, 30%) while the average intensity values need to be decreased (for example, 105) to include more peaks. After manually checking peaks, components with good shapes are selected and searched for in the human metabolome database. The results are shown in Table 2. Table 2A lists the results from data collected in positive mode, while Table 2B shows the results from negative mode. Some of the metabolites identified here overlap with the metabolites in the targeted list, such as glutathione, proline and so on, but meanwhile, additional metabolites absent from the targeted list are explored, such as methyglyoxal, which can be derived from glycolysis, and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, which is detected in positive with a retention time of 3.2 min, which is a reasonable retention for phospholipids on an amide column. A protocol on untargeted metabolite database searching has been previously reported20.

Figure 1. Examples of LC-MS chromatography peaks. Here, the reconstructed chromatography is generated with a mass window of 10 ppm (m/z ± 5 ppm). The X axis shows the retention time, while the Y axis shows the relative intensity, and the peak intensity is listed above every metabolite. A shows peaks detected from positive mode, while B shows peaks from negative mode. Click here to view larger image.
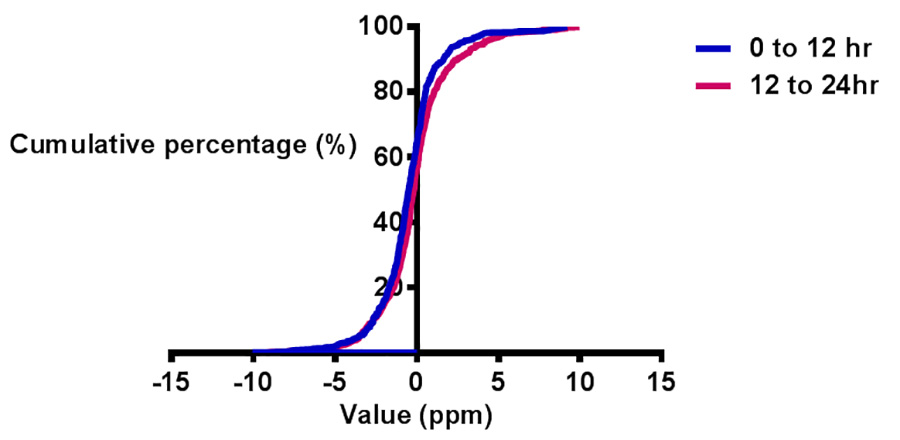
Figure 2. Evaluation of low mass range calibration. The Y axis is the cumulative percentage of metabolites with mass detection error within 5 ppm. The X axis is the mass error range in ppm. Blue and red curves represent 0-12 hr and 12-24 hr, respectively. Click here to view larger image.
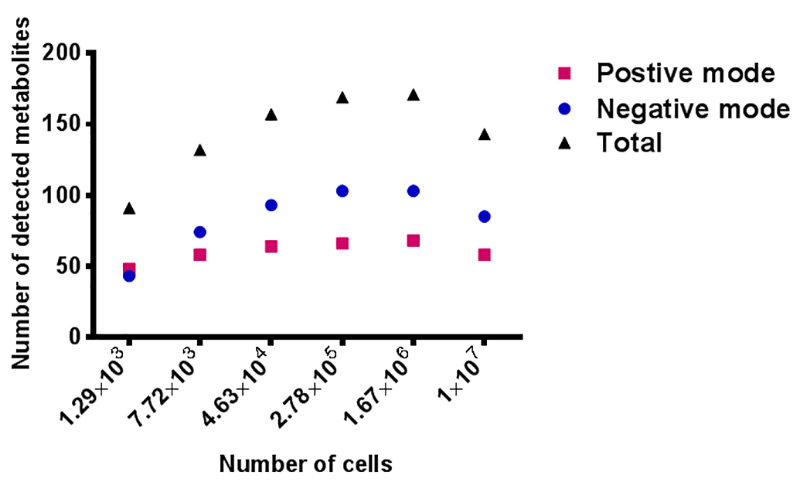
Figure 3. Evaluation of sample amount – number of targeted metabolites detected versus number of HCT 8 cells. Red squares represent metabolites detected in positive mode, blue circles mean metabolites measured in negative mode, and the black triangles are the total numbers of metabolites from both positive and negative mode. The X axis shows the number of HCT 8 cells. Click here to view larger image.
| Standards | M/Z, positive mode | M/Z, negative mode | Neutral formula | Neutral mass |
| n-Butylamine | 74.096425 | NA | C4H11N | 73.089149 |
| Caffeine fragment | 138.066188 | NA | C6H8N3O | 137.058912 |
| Caffeine | 195.087652 | NA | C8H11N4O2 | 194.080376 |
| Diazinon | 305.108329 | NA | C12H20N2O3PS | 304.101053 |
| MRFA peptide | 524.264966 | NA | C23H37N7O5S | 523.25769 |
| Fluoroacetate | N/A | 77.004432 | C2H3FO2 | 78.011708 |
| Sulfate | N/A | 96.960106 | H2SO4 | 97.967382 |
| Homovanillic acid | N/A | 181.050634 | C9H10O4 | 182.05791 |
| Dodecyl sulfate | N/A | 265.147906 | C12H26SO4 | 266.155182 |
| Taurocholate | N/A | 514.2844 | C26H45NO7S | 515.291676 |
Table 1. Low mass range calibration standards and their exact m/z. The formula shown here is corresponding to the neutral form formula, and m/z is the neutral mass plus or minus a proton.
| CSID | Name | Formula | Monoisotopic Mass | Search Mass | Error (ppm) | R.T. (min) |
| 234 | beta-Alanine | C3H7NO2 | 89.04800 | 89.04805 | 0.62 | 8.08 |
| 1057 | Sarcosine | C3H7NO2 | 89.04768 | 89.04805 | 4.22 | 8.08 |
| 5735 | alanine | C3H7NO2 | 89.04768 | 89.04805 | 4.22 | 8.08 |
| 568 | Creatinine | C4H7N3O | 113.05900 | 113.05889 | 1.00 | 4.41 |
| 128566 | Proline | C5H9NO2 | 115.06333 | 115.06338 | 0.41 | 7.54 |
| 6050 | L-(+)-Valine | C5H11NO2 | 117.07898 | 117.07896 | 0.18 | 7.42 |
| 7762 | Amyl nitrite I | C5H11NO2 | 117.07898 | 117.07896 | 0.18 | 7.42 |
| 135 | 5-amino valeric acid | C5H11NO2 | 117.07900 | 117.07896 | 0.38 | 7.42 |
| 242 | trimethylglycine | C5H11NO2 | 117.07900 | 117.07896 | 0.38 | 7.42 |
| 911 | Niacinamide | C6H6N2O | 122.04800 | 122.04793 | 0.55 | 2.60 |
| 1091 | Taurin | C2H7NO3S | 125.01466 | 125.01469 | 0.20 | 7.61 |
| 1030 | Pyrroline hydroxycarboxylic acid | C5H7NO3 | 129.04259 | 129.04259 | 0.02 | 8.51 |
| 7127 | PCA | C5H7NO3 | 129.04259 | 129.04259 | 0.02 | 8.51 |
| 90657 | N-Acryloylglycine | C5H7NO3 | 129.04259 | 129.04259 | 0.02 | 8.51 |
| 388752 | 5-Oxo-D-prolin | C5H7NO3 | 129.04259 | 129.04259 | 0.02 | 8.51 |
| 389257 | 3-Hydroxy-3,4-dihydro-2H-pyrrole-5-carboxylic acid | C5H7NO3 | 129.04259 | 129.04259 | 0.02 | 8.51 |
| 8031176 | Pyrrolidonecarboxylic acid | C5H7NO3 | 129.04259 | 129.04259 | 0.03 | 8.51 |
| 5605 | Hydroxyproline | C5H9NO3 | 131.05824 | 131.05901 | 5.83 | 8.08 |
| 7068 | N-Acetylalanin | C5H9NO3 | 131.05824 | 131.05901 | 5.83 | 8.08 |
| 79449 | Ac-Ala-OH | C5H9NO3 | 131.05824 | 131.05901 | 5.83 | 8.08 |
| 89122 | Ethylformylglycine | C5H9NO3 | 131.05824 | 131.05901 | 5.83 | 8.08 |
| 167744 | l-Glutamic-gamma-semialdehyde | C5H9NO3 | 131.05824 | 131.05901 | 5.83 | 8.08 |
| 388519 | 5-Amino-2-oxopentanoic acid | C5H9NO3 | 131.05824 | 131.05901 | 5.83 | 8.08 |
| 134 | Aminolevulinic acid | C5H9NO3 | 131.05800 | 131.05901 | 7.70 | 8.08 |
| 9312313 | 3-Hydroxy-L-proline | C5H9NO3 | 131.05800 | 131.05901 | 7.70 | 8.08 |
| 566 | Creatine | C4H9N3O2 | 131.06900 | 131.06905 | 0.36 | 8.08 |
| 5880 | L-(+)-Leucine | C6H13NO2 | 131.09464 | 131.09455 | 0.68 | 6.96 |
| 6067 | L-(+)-Isoleucine | C6H13NO2 | 131.09464 | 131.09455 | 0.68 | 6.96 |
| 19964 | L-Norleucine | C6H13NO2 | 131.09464 | 131.09455 | 0.68 | 6.96 |
| 388796 | beta-Leucine | C6H13NO2 | 131.09464 | 131.09455 | 0.68 | 6.96 |
| 548 | Aminocaproic acid | C6H13NO2 | 131.09500 | 131.09455 | 3.48 | 6.96 |
| 6031 | L-(-)-Asparagine | C4H8N2O3 | 132.05350 | 132.05348 | 0.10 | 8.31 |
| 109 | Ureidopropionic acid | C4H8N2O3 | 132.05299 | 132.05348 | 3.71 | 8.31 |
| 6026 | L-Ornithine | C5H12N2O2 | 132.08987 | 132.08988 | 0.02 | 10.37 |
| 64236 | D-Ornithine | C5H12N2O2 | 132.08987 | 132.08988 | 0.02 | 10.37 |
| 5746 | Glutamine | C5H10N2O3 | 146.06914 | 146.06900 | 0.93 | 8.25 |
| 128633 | D-Glutamine | C5H10N2O3 | 146.06914 | 146.06900 | 0.93 | 8.25 |
| 141172 | Ureidoisobutyric acid | C5H10N2O3 | 146.06914 | 146.06900 | 0.93 | 8.25 |
| 21436 | N-Methyl-<scp>D</scp>-aspartic acid | C5H9NO4 | 147.05316 | 147.05300 | 1.13 | 8.06 |
| 21814 | D-(-)-Glutamic acid | C5H9NO4 | 147.05316 | 147.05300 | 1.13 | 8.06 |
| 30572 | L-(+)-Glutamic acid | C5H9NO4 | 147.05316 | 147.05300 | 1.13 | 8.06 |
| 58744 | N-Acetyl-L-serine | C5H9NO4 | 147.05316 | 147.05300 | 1.13 | 8.06 |
| 5907 | L-(-)-methionine | C5H11NO2S | 149.05106 | 149.05095 | 0.70 | 7.39 |
| 6038 | Histidine | C6H9N3O2 | 155.06947 | 155.06940 | 0.48 | 8.33 |
| 5910 | L-(-)-Phenylalanine | C9H11NO2 | 165.07898 | 165.07887 | 0.63 | 6.60 |
| 1025 | Pyridoxine | C8H11NO3 | 169.07390 | 169.07376 | 0.80 | 3.24 |
| 4463 | Oxidopamine [USAN:INN] | C8H11NO3 | 169.07390 | 169.07376 | 0.80 | 3.24 |
| 102750 | 5-(2-aminoethyl)-Pyrogallol | C8H11NO3 | 169.07390 | 169.07376 | 0.80 | 3.24 |
| 388394 | Norepinephrine | C8H11NO3 | 169.07390 | 169.07376 | 0.80 | 3.24 |
| 6082 | L-(+)-Arginine | C6H14N4O2 | 174.11168 | 174.11144 | 1.39 | 10.77 |
| 64224 | D-Arg | C6H14N4O2 | 174.11168 | 174.11144 | 1.39 | 10.77 |
| 780 | heteroauxin | C10H9NO2 | 175.06300 | 175.06304 | 0.18 | 2.32 |
| 67261 | Indole-3-acetaldehyde, 5-hydroxy- | C10H9NO2 | 175.06332 | 175.06304 | 1.65 | 2.32 |
| 3574185 | INDOLE-2-ACETIC ACID | C10H9NO2 | 175.06332 | 175.06304 | 1.65 | 2.32 |
| 5833 | L-(-)-Tyrosine | C9H11NO3 | 181.07390 | 181.07378 | 0.66 | 7.48 |
| 389285 | 3-Amino-3-(4-hydroxyphenyl)propanoic acid | C9H11NO3 | 181.07390 | 181.07378 | 0.66 | 7.48 |
| 13628311 | L-threo-3-phenylserine | C9H11NO3 | 181.07390 | 181.07378 | 0.66 | 7.48 |
| 425 | 4-hydroxy-4-(3-pyridyl)butanoic acid | C9H11NO3 | 181.07401 | 181.07378 | 1.25 | 7.48 |
| 13899 | 3-(1H-Indol-3-yl)acrylic acid | C11H9NO2 | 187.06332 | 187.06330 | 0.15 | 6.44 |
| 10607876 | Indoleacrylic acid | C11H9NO2 | 187.06332 | 187.06330 | 0.15 | 6.44 |
| 389120 | N6,N6,N6-Trimethyl-L-lysine | C9H20N2O2 | 188.15248 | 188.15221 | 1.45 | 10.87 |
| 388321 | 5"-S-Methyl-5"-thioadenosine | C11H15N5O3S | 297.08957 | 297.08898 | 2.00 | 2.56 |
| 144 | 9-(5-s-methyl-5-thiopentofuranosyl)-9h-purin-6-amine | C11H15N5O3S | 297.09000 | 297.08898 | 3.43 | 2.56 |
| 111188 | Glutathione | C10H17N3O6S | 307.08380 | 307.08345 | 1.14 | 8.02 |
Table 2A.
| CSID | Name | Formula | Monoisotopic Mass | Search Mass | Error (ppm) | R.T. (min) |
| 857 | Methylglyoxal | C3H4O2 | 72.02100 | 72.02108 | 1.03 | 7.70 |
| 1057 | Sarcosine | C3H7NO2 | 89.04768 | 89.04747 | 2.33 | 8.19 |
| 5735 | alanine | C3H7NO2 | 89.04768 | 89.04747 | 2.33 | 8.19 |
| 234 | beta-Alanine | C3H7NO2 | 89.04800 | 89.04747 | 5.93 | 8.19 |
| 55423 | R-lactic acid | C3H6O3 | 90.03169 | 90.03143 | 2.91 | 5.12 |
| 61460 | Hydroxypropionic acid | C3H6O3 | 90.03169 | 90.03143 | 2.91 | 5.12 |
| 96860 | L-(+)-lactic acid | C3H6O3 | 90.03169 | 90.03143 | 2.91 | 5.12 |
| 592 | Lactic acid | C3H6O3 | 90.03200 | 90.03143 | 6.29 | 5.12 |
| 650 | Dihydroxyacetone | C3H6O3 | 90.03200 | 90.03143 | 6.29 | 5.12 |
| 731 | Glyceraldehyde | C3H6O3 | 90.03200 | 90.03143 | 6.29 | 5.12 |
| 1086 | Sulfuric acid | H2O4S | 97.96738 | 97.96683 | 5.63 | 8.13 |
| 128566 | Proline | C5H9NO2 | 115.06333 | 115.06302 | 2.67 | 7.78 |
| 1078 | Succinic acid | C4H6O4 | 118.02661 | 118.02630 | 2.66 | 7.72 |
| 466979 | Erythrono-1,4-lactone | C4H6O4 | 118.02661 | 118.02630 | 2.66 | 7.72 |
| 4483398 | D-Erythronic g-lactone | C4H6O4 | 118.02661 | 118.02630 | 2.66 | 7.72 |
| 473 | Methylmalonic acid | C4H6O4 | 118.02700 | 118.02630 | 5.96 | 7.72 |
| 8527138 | (3S,4R)-3,4-Dihydroxydihydrofuran-2(3H)-one | C4H6O4 | 118.02700 | 118.02630 | 5.96 | 7.72 |
| 140384 | 2-ketocaproic acid | C6H10O3 | 130.06299 | 130.06270 | 2.24 | 2.35 |
| 164251 | Methyloxovaleric acid | C6H10O3 | 130.06299 | 130.06270 | 2.24 | 2.35 |
| 388419 | (3S)-3-Methyl-2-oxopentanoic acid | C6H10O3 | 130.06299 | 130.06270 | 2.24 | 2.35 |
| 15642233 | Ketoleucine | C6H10O3 | 130.06299 | 130.06270 | 2.24 | 2.35 |
| 46 | a-Oxo-b-methylvaleric acid | C6H10O3 | 130.06300 | 130.06270 | 2.36 | 2.35 |
| 69 | Alpha-ketoisocaproic acid | C6H10O3 | 130.06300 | 130.06270 | 2.36 | 2.35 |
| 134 | Aminolevulinic acid | C5H9NO3 | 131.05800 | 131.05795 | 0.36 | 8.19 |
| 9312313 | 3-Hydroxy-L-proline | C5H9NO3 | 131.05800 | 131.05795 | 0.36 | 8.19 |
| 5605 | HYDROXYPROLINE | C5H9NO3 | 131.05824 | 131.05795 | 2.23 | 8.19 |
| 7068 | N-Acetylalanin | C5H9NO3 | 131.05824 | 131.05795 | 2.23 | 8.19 |
| 79449 | Ac-Ala-OH | C5H9NO3 | 131.05824 | 131.05795 | 2.23 | 8.19 |
| 89122 | Ethylformylglycine | C5H9NO3 | 131.05824 | 131.05795 | 2.23 | 8.19 |
| 167744 | l-Glutamic-gamma-semialdehyde | C5H9NO3 | 131.05824 | 131.05795 | 2.23 | 8.19 |
| 388519 | 5-Amino-2-oxopentanoic acid | C5H9NO3 | 131.05824 | 131.05795 | 2.23 | 8.19 |
| 5880 | L-(+)-Leucine | C6H13NO2 | 131.09464 | 131.09419 | 3.39 | 7.09 |
| 6067 | L-(+)-Isoleucine | C6H13NO2 | 131.09464 | 131.09419 | 3.39 | 7.09 |
| 19964 | L-Norleucine | C6H13NO2 | 131.09464 | 131.09419 | 3.39 | 7.09 |
| 388796 | beta-Leucine | C6H13NO2 | 131.09464 | 131.09419 | 3.39 | 7.09 |
| 548 | Aminocaproic acid | C6H13NO2 | 131.09500 | 131.09419 | 6.18 | 7.09 |
| 109 | Ureidopropionic acid | C4H8N2O3 | 132.05299 | 132.05321 | 1.60 | 8.28 |
| 6031 | L-(-)-Asparagine | C4H8N2O3 | 132.05350 | 132.05321 | 2.21 | 8.28 |
| 6026 | L-Ornithine | C5H12N2O2 | 132.08987 | 132.08961 | 1.98 | 10.36 |
| 64236 | D-Ornithine | C5H12N2O2 | 132.08987 | 132.08961 | 1.98 | 10.36 |
| 193317 | L-(−)-Malic acid | C4H6O5 | 134.02153 | 134.02130 | 1.72 | 7.95 |
| 510 | (±)-Malic Acid | C4H6O5 | 134.02200 | 134.02130 | 5.25 | 7.95 |
| 133224 | threonic acid | C4H8O5 | 136.03717 | 136.03688 | 2.14 | 7.70 |
| 388628 | 2,3,4-Trihydroxybutanoic acid | C4H8O5 | 136.03717 | 136.03688 | 2.14 | 7.70 |
| 2061231 | DL-erythronic acid | C4H8O5 | 136.03717 | 136.03688 | 2.14 | 7.70 |
| 21436 | N-Methyl-<scp>D</scp>-aspartic acid | C5H9NO4 | 147.05316 | 147.05299 | 1.16 | 8.05 |
| 21814 | D-(-)-Glutamic acid | C5H9NO4 | 147.05316 | 147.05299 | 1.16 | 8.05 |
| 30572 | L-(+)-Glutamic acid | C5H9NO4 | 147.05316 | 147.05299 | 1.16 | 8.05 |
| 58744 | N-Acetyl-L-serine | C5H9NO4 | 147.05316 | 147.05299 | 1.16 | 8.05 |
| 6038 | Histidine | C6H9N3O2 | 155.06947 | 155.06930 | 1.09 | 8.36 |
| 199 | Allantoin | C4H6N4O3 | 158.04401 | 158.04387 | 0.88 | 4.76 |
| 6082 | L-(+)-Arginine | C6H14N4O2 | 174.11168 | 174.11154 | 0.80 | 10.76 |
| 64224 | D-Arg | C6H14N4O2 | 174.11168 | 174.11154 | 0.80 | 10.76 |
| 58576 | N-Acetyl-L-Aspartic acid | C6H9NO5 | 175.04807 | 175.04803 | 0.18 | 7.87 |
| 996 | Pyrophosphoric Acid | H4O7P2 | 177.94299 | 177.94331 | 1.79 | 8.42 |
| 5589 | Glucose | C6H12O6 | 180.06339 | 180.06346 | 0.41 | 7.53 |
| 17893 | Mannose | C6H12O6 | 180.06339 | 180.06346 | 0.41 | 7.53 |
| 58238 | .beta.-D-Glucopyranose | C6H12O6 | 180.06339 | 180.06346 | 0.41 | 7.53 |
| 71358 | .alpha.-D-Glucopyranose | C6H12O6 | 180.06339 | 180.06346 | 0.41 | 7.53 |
| 134838 | 3-deoxy-arabino-hexonic acid | C6H12O6 | 180.06339 | 180.06346 | 0.41 | 7.53 |
| 388332 | L-Sorbopyranose | C6H12O6 | 180.06339 | 180.06346 | 0.41 | 7.53 |
| 388476 | beta-D-galactopyranose | C6H12O6 | 180.06339 | 180.06346 | 0.41 | 7.53 |
| 388480 | .alpha.-D-Galactopyranose | C6H12O6 | 180.06339 | 180.06346 | 0.41 | 7.53 |
| 388775 | beta-D-Fructofuranose | C6H12O6 | 180.06339 | 180.06346 | 0.41 | 7.53 |
| 10239179 | Inositol | C6H12O6 | 180.06339 | 180.06346 | 0.41 | 7.53 |
| 16736992 | Cis-inositol | C6H12O6 | 180.06339 | 180.06346 | 0.41 | 7.53 |
| 17216070 | allose | C6H12O6 | 180.06339 | 180.06346 | 0.41 | 7.53 |
| 17216093 | L-Sorbose | C6H12O6 | 180.06339 | 180.06346 | 0.41 | 7.53 |
| 201 | hexopyranose | C6H12O6 | 180.06300 | 180.06346 | 2.53 | 7.53 |
| 868 | 1,2,3,4,5,6-cyclohexanhexol | C6H12O6 | 180.06300 | 180.06346 | 2.53 | 7.53 |
| 2068 | Theophylline | C7H8N4O2 | 180.06473 | 180.06346 | 7.04 | 7.53 |
| 4525 | 1,7-dimethyl-Xanthine | C7H8N4O2 | 180.06473 | 180.06346 | 7.04 | 7.53 |
| 5236 | Theobromine | C7H8N4O2 | 180.06473 | 180.06346 | 7.04 | 7.53 |
| 1161 | isocitric acid | C6H8O7 | 192.02701 | 192.02704 | 0.15 | 8.18 |
| 305 | Citric acid | C6H8O7 | 192.02699 | 192.02704 | 0.23 | 8.18 |
| 963 | pantothenic acid | C9H17NO5 | 219.11067 | 219.11049 | 0.84 | 6.72 |
| 6361 | D-pantothenic acid | C9H17NO5 | 219.11067 | 219.11049 | 0.84 | 6.72 |
| 960 | palmitic acid | C16H32O2 | 256.23999 | 256.24000 | 0.05 | 1.73 |
| 111188 | Glutathione | C10H17N3O6S | 307.08380 | 307.08339 | 1.35 | 8.03 |
| 388337 | N-Acetylneuraminic acid | C11H19NO9 | 309.10599 | 309.10585 | 0.45 | 7.43 |
| 392681 | N-Acetyl-alpha-neuraminic acid | C11H19NO9 | 309.10599 | 309.10585 | 0.45 | 7.43 |
| 392810 | Sialic acid Neu5Ac | C11H19NO9 | 309.10599 | 309.10585 | 0.45 | 7.43 |
Table 2B.
Table 2. List of untargeted metabolites detected in HCT 8 cells (2.78 x 105 cells equivalence). Table 2A and 2B include components extraction information: retention time, m/z and mass error, and meanwhile, database search results: Chemspider ID (CSID), name, formula, and so on. Here the samples analyzed are equal to metabolites extracted from 2.78 x 105 cells, and the intensity threshold is 1 x 107 to avoid tedious result for demonstration aim.
Discussion
The most critical steps for successful metabolite profiling in cells using this protocol are: 1) controlling the growth medium and careful extraction of the cells; 2) adjusting the LC method based on MS method setup to ensure there are enough (usually at least 10) data points across a peak for quantitation; 3) doing a low mass calibration before running samples; 4) injecting no more than 5 ml to avoid retention time shifting and peak broadening caused by water; and 5) preparing and running samples for comparison in the same batch to minimize batch effects.
The standards (Table 1) chosen here for low mass range calibration are interchangeable. Any known compound with an m/z that falls into the mass scan range, is well behaved in a H-ESI source, and is soluble in water, methanol, or acetonitrile are reasonable candidates for calibration standards. It is highly recommended to store all calibration solutions at 4 °C to stabilize caffeine and also to minimize the evaporation of methanol or acetonitrile in the calibration solution so that the calibration performance can be more reproducible. Compared to a regular mass range, m/z from 150 to 2,000, low mass range calibration needs to be done more frequently, at least once every two days.
This workflow, from extraction solvent, reconstitution solvent, LC mobile phase, to low mass range calibration and MS scan range has been optimized to measure polar metabolites. This includes amino acids, acetyl amino acids, glycolysis pathway intermediates, nucleosides, TCA cycle intermediates, some one-carbon metabolism pathway intermediates and so on. However, modifications of this protocol for other classes of metabolites, such as Coenzyme A (CoA) species, folates, phospholipids are possible. For example, CoAs are more stable in acidic conditions, so the addition of an acid to 80% methanol/water will be helpful to improve CoA sensitivity. Also, CoAs and lipids tend to have much large molecular weights, thus the m/z scan range needs to be adjusted from 60-900 to the proper range which will cover those metabolites.
Even though some of the untargeted component database search results overlap with the targeted list, it is still of importance to build this targeted list based on the research priority. Since the metabolites in the targeted list are lower than the average intensity threshold, information on these metabolites will be removed during processing. The targeted list includes the retention time information, which gives us higher confidence for metabolite identification and quantitation. One further advantage with the QE-MS setup is that tandem mass spectrometry can allow for further identification of metabolites.
One issue associated with this workflow is that the H-ESI needle insert is sensitive to the salt content of the samples, as the sensitivity will be greatly compromised if there are high amounts of non-volatile salts. Therefore, minimizing salt content from samples, and routine cleaning of the column and the H-ESI needle insert will be helpful to ensure good quality data and to increase the column’s lifetime.
In summary, this protocol employs LC-QE-MS to successfully analyze polar metabolites from cultured cells, with minimal sample preparation steps and rapid data acquisition. Small modifications in sample preparation can be carried out to obtain data from other biological sources such as serum and tissue. For example, since pure methanol can be added to liquid serum added to make a final methanol concentration to 80% for polar metabolites extraction. For tissue samples, rigorous stirring and mixing is required to achieve better extraction efficiency. Usually 10 ml serum or 1 mg tissue is sufficient for metabolites analysis. The raw data can be analyzed both in targeted mode, if there are known metabolites in the samples, and in an un-targeted way followed by HRMS database searching. HRMS based metabolomics is still in its early stages. For future advances, the experimental techniques can be further optimized, additional metabolite HRMS information and MS/MS fragmentation patterns will be helpful and relevant algorithms, such as peak alignment, peak integration, isotope clustering and so on, can improve the efficiency and accuracy of data processing. Ultimately, however, many of the questions that our lab addresses in metabolism are limited by careful interpretation of data after processing. With these large-scale metabolomics techniques, we are often limited by our interpretation of the data and evaluation of hypotheses generated. Therefore, all metabolomics experiments need be formulated around specific questions.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors would like to acknowledge Detlef Schumann, Jennifer Sutton (Thermo Fisher Scientific) and Nathaniel Snyder (University of Pennsylvania) for valuable discussions on mass calibration and data processing. Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number R00CA168997. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Materials
| Positive calibration mix | Thermo Scientific | #88323 | It is light sensitive. Store at 4 °C |
| Negative calibration mix | Thermo Scientific | #88324 | Store at 4 °C |
| Diazinon | Sant Cruz Biotechnology | #C0413 | It causes eyes irritation, so work in hood. Store at 4 °C |
| H-ESI needle insert | Fisher Scientific | #1303200 | This could be replaced or cleaned with 5 % Formic acid/water (remove rubber ring) if clogged. |
| Xbridge amide column | Waters | #186004860 | Guard column is recommend to increase column lifetime. |
References
- Fiehn, O. Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comparative and Functional Genomics. 2 (3), 155-168 (2001).
- Mulleder, M., et al. A prototrophic deletion mutant collection for yeast metabolomics and systems biology. Nature Biotechnology. 30 (12), 1176-1178 (2012).
- Patel, V. R., Eckel-Mahan, K., Sassone-Corsi, P., Baldi, P. CircadiOmics: Integrating circadian genomics, transcriptomics, proteomics and metabolomics. Nature Methods. 9 (8), 772-773 (2012).
- Ideker, T., et al. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 292 (5518), 929-934 (2001).
- Jonsson, P., et al. A strategy for identifying differences in large series of metabolomic samples analyzed by GC/MS. Analytical Chemistry. 76 (6), 1738-1745 (2004).
- Jonsson, P., et al. High-throughput data analysis for detecting and identifying differences between samples in GC/MS-based metabolomic analyses. Analytical Chemistry. 77 (17), 5635-5642 (2005).
- Weljie, A. M., Newton, J., Mercier, P., Carlson, E., Slupsky, C. M. Targeted profiling: Quantitative analysis of 1H NMR metabolomics data. Analytical Chemistry. 78 (13), 4430-4442 (2006).
- Wiklund, S., et al. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Analytical Chemistry. 80 (1), 115-122 (2008).
- Xia, J., Bjorndahl, T. C., Tang, P., Wishart, D. S. MetaboMiner–semi-automated identification of metabolites from 2D NMR spectra of complex biofluids. BMC Bioinformatics. 9, 507 (2008).
- Lu, D., et al. 13C NMR isotopomer analysis reveals a connection between pyruvate cycling and glucose-stimulated insulin secretion (GSIS). Proceedings of the National Academy of Sciences of the United States of America. 99 (5), 2708-2713 (2002).
- Shen, J., et al. Determination of the rate of the glutamate/glutamine cycle in the human brain b. in vivo 13C NMR. Proceedings of the National Academy of Sciences of the United States of America. 96 (14), 8235-8240 (1999).
- Kitteringham, N. R., Jenkins, R. E., Lane, C. S., Elliott, V. L., Park, B. K. Multiple reaction monitoring for quantitative biomarker analysis in proteomics and metabolomics. Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences. 877 (13), 1229-1239 (2009).
- Locasale, J. W., et al. Metabolomics of human cerebrospinal fluid identifies signatures of malignant glioma. Molecular & Cellular Proteomics : MCP. 11 (6), 10-1074 (2012).
- Wolf-Yadlin, A., Hautaniemi, S., Lauffenburger, D. A., White, F. M. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proceedings of the National Academy of Sciences of the United States of America. 104 (14), 5860-5865 (2007).
- Yuan, M., Breitkopf, S. B., Yang, X., Asara, J. M. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nature Protocols. 7 (5), 872-881 (2012).
- Ramanathan, R., et al. It is time for a paradigm shift in drug discovery bioanalysis: From SRM to HRMS. Journal of Mass Spectrometry : JM. 46 (6), 595-601 (2011).
- Lu, W., Clasquin, M. F., Melamud, E., Amador-Noguez, D., Caudy, A. A., Rabinowitz, J. D. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Analytical Chemistry. 82 (8), 3212-3221 (2010).
- Michalski, A., et al. Ultra high resolution linear ion trap orbitrap mass spectrometer (orbitrap elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes. Molecular & Cellular Proteomics : MCP. 11 (3), 10-1074 (2012).
- Michalski, A., et al. Mass spectrometry-based proteomics using Q exactive, a high-performance benchtop quadrupole orbitrap mass spectrometer. Molecular & Cellular Proteomics : MCP. 10 (9), (2011).
- Snyder, N. W., Khezam, M., Mesaros, C. A., Worth, A., Blair, I. A. Untargeted Metabolomics from Biological Sources Using Ultraperformance Liquid Chromatography-High Resolution Mass Spectrometry. UPLC-HRMS). J. Vis. Exp. (75), (2013).
- Gika, H. G., Theodoridis, G. A., Wingate, J. E., Wilson, I. D. Within-day reproducibility of an HPLC-MS-based method for metabonomic analysis: Application to human urine. Journal of Proteome Research. 6 (8), 3291-3303 (2007).
- Want, E. J., et al. Global metabolic profiling procedures for urine using UPLC-MS. Nature Protocols. 5 (6), 1005-1018 (2010).

