Formation of Biomembrane Microarrays with a Squeegee-based Assembly Method
Summary
Supported lipid bilayers and natural membrane particles are convenient systems that can approximate the properties of cell membranes and be incorporated in a variety of analytical strategies. Here we demonstrate a method for preparing microarrays composed of supported lipid bilayer-coated SiO2 beads, phospholipid vesicles or natural membrane particles.
Abstract
Lipid bilayer membranes form the plasma membranes of cells and define the boundaries of subcellular organelles. In nature, these membranes are heterogeneous mixtures of many types of lipids, contain membrane-bound proteins and are decorated with carbohydrates. In some experiments, it is desirable to decouple the biophysical or biochemical properties of the lipid bilayer from those of the natural membrane. Such cases call for the use of model systems such as giant vesicles, liposomes or supported lipid bilayers (SLBs). Arrays of SLBs are particularly attractive for sensing applications and mimicking cell-cell interactions. Here we describe a new method for forming SLB arrays. Submicron-diameter SiO2 beads are first coated with lipid bilayers to form spherical SLBs (SSLBs). The beads are then deposited into an array of micro-fabricated submicron-diameter microwells. The preparation technique uses a “squeegee” to clean the substrate surface, while leaving behind SSLBs that have settled into microwells. This method requires no chemical modification of the microwell substrate, nor any particular targeting ligands on the SSLB. Microwells are occupied by single beads because the well diameter is tuned to be just larger than the bead diameter. Typically, more 75% of the wells are occupied, while the rest remain empty. In buffer SSLB arrays display long-term stability of greater than one week. Multiple types of SSLBs can be placed in a single array by serial deposition, and the arrays can be used for sensing, which we demonstrate by characterizing the interaction of cholera toxin with ganglioside GM1. We also show that phospholipid vesicles without the bead supports and biomembranes from cellular sources can be arrayed with the same method and cell-specific membrane lipids can be identified.
Introduction
Lipid bilayer membranes are essential structures in nature. Cellular plasma membranes and organelle membranes are composed of lipid bilayers that incorporate a number of molecules that are necessary for life. Many life-sustaining processes occur on the surface of cells or are mediated by molecules associated with lipid-bilayer membranes. In fact, many pharmaceuticals target processes or molecules are found on or in membranes1,2. It is therefore necessary to analytically investigate processes, such as chemical reactions or noncovalent binding events that occur on membrane surfaces. Because natural membranes can be difficult to isolate and/or interface with sensors, many researchers employ simplified model membranes to carry out analytical studies. A number of model membrane systems are described in the literature, ranging from giant vesicles that can be tens to hundreds of microns in diameter to liposomes with nanoscale dimensions3,4. Alternatively, planar lipid bilayers deposited on solid supports, i.e., supported lipid bilayers (SLBs), can be formed on a number of different surfaces and have been widely used in biophysical, biochemical, and analytical applications5. Coupling SLBs with electrical or optical materials enables investigation of membrane biochemistry and biophysics through the use of different analytical techniques. Fluorescence microscopy6, electrochemistry7, optical spectroscopy8, scanning probe microscopy9, surface plasmon resonance10, and mass spectrometry11 have all been employed to study the structure and properties of SLBs.
SLB arrays offer additional versatility in the design of sensors for multiplex assays12,13. Other applications use SLB arrays to mimic the junction that forms between immune cells14. Preparation methods for SLB arrays have varied from microfluidic approaches15 to those that employ physical barriers between adjacent SLB patches.16 Other groups have used printing methods17, photochemical patterning18 and various nanoengineering approaches19 to create SLB arrays.
In this paper and accompanying video we demonstrate a method for forming SLB arrays by depositing SLB-coated SiO2 beads into ordered arrays of microwells20. We refer to the SLB-coated SiO2 beads as spherical supported lipid bilayers (SSLBs). This technique is an extension of earlier work that created arrays of phospholipid vesicles and biomembranes derived from natural sources21, from which we also show example results. Other methods for arraying biomembrane particles or vesicles have relied on patterns of specific targeting ligands on surfaces that associate with complementary ligands contained on the vesicle surface. Examples include biotin-avidin association22,23 and DNA hybridization schemes24. Our approach only requires a microwell array with no targeting or recognition moieties necessary. The size of SSLBs is defined by the diameter of the SiO2 bead supports, which have low poly-dispersity. By tuning the microwell diameter to just larger than the SSLB diameter, only a single SSLB settles into each microwell. A poly(dimethylsiloxane) (PDMS) squeegee then removes from the surface all SSLBs that are not immobilized in microwells. The microwells and resultant SSLB arrays have high density (~105 SSLBs/mm2) with 3 µm center-to-center spacing and hexagonal periodicity. By serially depositing SSLBs with different lipid compositions, it is possible to create multicomponent arrays with randomly positioned SSLBs. To demonstrate the sensing capability of SSLB arrays, we used the interaction of cholera toxin (CTx) with a ganglioside (GM1) incorporated into the SSLBs. With natural membrane particles, we were able to detect cell-specific lipids in multicomponent arrays containing membrane material from two different cell types.
Protocol
1. Microfabrication of Microwell Array Substrate
- Start with a 4 inch silicon wafer with 100 nm of thermally grown oxide.
- Spin SPR-955 0.7 photoresist on the wafer at 4,000 rpm for 30 sec.
- Bake on a hotplate at 115 °C for 90 sec.
- Expose photoresist.
- Use a mask that will create 1 µm holes arranged in a hexagonal array with a 3 µm period where the array covers a 2 mm x 2 mm area.
- Expose wafer in an i-line stepper using a step size of 6 mm and an exposure of 200 mJ/cm2.
- Bake on a hotplate at 115 °C for 90 sec.
- Develop using a spin developer to deposit 2 puddles of CD-26 on the wafer for a total development time of 90 sec.
- Rinse thoroughly with ultrapure DI water and dry with N2.
- Etch the oxide in a reactive-ion etcher for 6 min with 50 sccm of Ar, 25 sccm of CF4, and 50 sccm of CHF3 flowing at a pressure of 75 mTorr.
- Etch silicon in an ICP deep-trench reactive-ion etcher for 2 min with 63 sccm of C4F8, 27 sccm of SF6, 40 sccm of Ar, and 10 sccm of O2 flowing at a pressure of 14 mTorr.
- Rinse in acetone, methanol, and isopropanol to remove resist.
- Place in a bath of H2SO4:H2O2 1:1 for 10 min to clean the surface.
- Rinse thoroughly with ultrapure DI water and dry with N2.
- Deposit 1,080 Å of Al2O3 by atomic layer deposition at 250 °C.
2. Preparation of Poly(dimethylsiloxane) Squeegee
- If possible, work in a cleanroom. Otherwise work in the cleanest environment possible.
- Obtain a plastic Petri dish (or other clean disposable container) with ≥13 mm sides.
- In that dish, pour out 5 g PDMS curing agent, followed by 50 g PDMS base agent (or anything with a 1:10 ratio that will mostly fill the Petri dish). Mix thoroughly with a disposable plastic pipette or rod.
- Remove bubbles by placing the mixed PDMS in a chamber with a vacuum line, applying vacuum until bubbles come out of mixture (approximately 30 min).
- If not already in the Petri dish, pour PDMS in Petri dish, avoiding creation of air bubbles.
- Cure overnight on a hotplate at >50 °C (higher heat for less time also works).
- With new/clean razor blade, carefully cut a rectangular piece approximately 2.5 x 2.5 cm, keeping top surface as clean as possible. Peel out with a clean forceps. Trim off one edge (down to 1.5 cm) by pressing down on razor blade in one motion, to make top edge as sharp as possible (this will be the edge used in the squeegee process).
- Clean with acetone, methanol, isopropyl alcohol, and ultrapure DI water, and then blow dry with clean nitrogen gas. Store in a clean Petri dish.
3. Preparation of Vesicles
- Collect lipid stock solutions of 25 mg/ml egg phosphatidylcholine (PC) and 1 mg/ml 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) ammonium salt (Rho-DPPE) in chloroform and a 0.5 mg/ml stock solution of monosialoganglioside GM1 in chloroform/methanol (2:1 v/v).
- In a small glass vial make a mixture that results in 97 mol% PC, 2 mol% GM1 and 1 mol% Rho-DPPE by adding 18.9 µl of 25 mg/ml PC, 39.6 µl of 0.5 mg/ml GM1 and 7.9 µl of 1 mg/ml Rho-DPPE using glass syringes.
- Note: The supplementary material for Nair et al.25 contains an excellent customizable Excel spreadsheet for calculating the amounts of lipids required to create vesicles of any desired composition.
- Place vial in a vacuum desiccator and remove the solvent by leaving the vial under vacuum for 6 hr.
- Make an aqueous solution of 0.1 M NaCl. Add 0.5 ml of the 0.1 M NaCl solution to the vial containing the dried lipid film.
- Allow the aqueous lipid mixture to rest overnight.
- Briefly vortex mix to create a vesicle suspension, then sonicate for 20 min in a bath sonicator at room temperature.
- Extrude the vesicle suspension through a polycarbonate membrane with 100 nm pore size. Pass the vesicle suspension through the filter 17x.
- Store the extruded vesicles in a glass vial at 4 °C.
4. Formation of Spherical Supported Lipid Bilayers
- Collect 700 nm diameter SiO2 beads. The beads are supplied in distilled water with a stock concentration of 1.4 x 1011 beads/ml.
- In a 1.5 ml Eppendorf tube prepare a 1 ml bead suspension with a concentration of 1.5 x 1010 beads/ml by adding 107 µl of bead stock solution to 893 µl of 0.1 M NaCl.
- Vortex the suspension to ensure it is well mixed.
- Centrifuge the suspension at 1,700 x g for 20 min then discard the supernatant. Resuspend the beads in 1 ml of 0.1 M NaCl. Repeat this step twice more to thoroughly wash the beads.
- To form the SSLBs, in a 1.5 ml Eppendorf tube add 25 µl of the SiO2 bead suspension to 200 µl of 1 mg/ml vesicle suspension. Vortex the mixture and let stand for 1 hr at room temperature. At this stage the SSLB concentration should be 1.7 x 109 SSLBs/ml.
- Centrifuge at 1,700 x g for 20 min to pellet the SSLBs. The pellet should appear pink due to the rupture of vesicles with Rho-DPPE on the beads.
- Discard the supernatant and resuspend in 225 µl phosphate buffered saline (PBS), pH = 7.4. Repeat Steps 4.6-4.7 two more times to remove any unruptured vesicles from the SSLB suspension.
5. Assembly of SSLB Array
- Cleave wafer of microwell arrays resulting in rectangular pieces with 4-6 microwell arrays per piece.
- Vortex mix the SSLB suspension, then place 10 µl of SSLB suspension on each microwell array. Lest rest for 1 hr to allow SSLBs to settle to the surface.
- Gently wash the microwell array chip with PBS from a wash bottle, then submerge in a PBS bath prepared in a shallow dish.
- While submerged, gently place the PDMS squeegee flush on the microwell array chip and gently slide it along the surface 5x to remove SSLBs that are not immobilized in microwells.
- Grip the microwell array chip with tweezers and gently shake in the PBS bath to remove any excess SSLBs.
- Quickly remove the chip from the squeegee bath and place in a fresh PBS bath until further use. Make sure the top surface of the chip remains wet to preserve the SSLBs.
Note: The assembly method described above can be used to create arrays of natural membrane particles, like myelin particles isolated from mouse brain, or phospholipid vesicles without the SiO2 bead supports. This work is described in detail in Wittenberg et al.21 and representative results are shown below.
6. Cholera Toxin Binding Assay
- Prepare a 2 mg/ml solution of bovine serum albumin (BSA) in PBS.
- Prepare a solution with desired concentration of Alexa 488-conjugated cholera toxin B-subunit in PBS with 2 mg/ml BSA.
- Remove the SSLB array chip from the PBS bath and wick away most of the PBS solution using a laboratory wipe. Leave just enough PBS on the chip to keep the SSLB array hydrated.
- To prevent nonspecific binding, add 200 µl of 2 mg/ml BSA solution to the SSLB array chip. Let rest for 1 hr in a humidified box.
- With a micropipette, remove 200 µl of solution from the top of the SSLB array chip.
- Add 200 µl of cholera toxin solution to the SSLB array chip and let rest for 1 hr in a humidified box.
- Gently wash the SSLB array chip with PBS from a wash bottle to remove any unbound cholera toxin.
- Wick away excess PBS using a laboratory wipe. Before imaging cover chip with a 24 mm x 40 mm cover slip.
- Image arrays with an upright microscope using filter sets appropriate for the fluorophores of interest.
- Analyze the fluorescence intensity of individual SSLBs in an array using the automated particle analysis function of ImageJ software.
- Compile mean fluorescence intensities from individual SSLBs into histograms that summarize the intensities of SSLBs found on a given array.
Representative Results
When SiO2 beads are mixed with a solution of vesicles composed of phospholipids, fluorescent lipids and other lipids such as gangliosides, the vesicles rupture on the SiO2 bead surfaces to form SSLBs, as shown schematically in Figure 1a. After washing the SSLBs, a drop of SSLB solution is placed on a microwell array, and the beads are allowed to settle to the surface. (Figure 1b1) This can also be done with a suspension of phospholipid vesicles or natural membrane particles, which are shown in Figure 1b2. A fluorescence image of phospholipid vesicles adsorbed to a microwell array substrate is shown in Figure 1b3. Then the PDMS squeegee is made to slide gently across the surface to remove any SSLBs (Figure 1c1), vesicles or natural membrane particles (Figure 1c2) that did not settle into microwells. This results in a SSLB array in which each microwell contains at most one SSLB (Figure 1d1) or an array where multiple vesicles or natural membrane particles are in each microwell (Figure 1d2). An image of a microarray composed of fluorescently labeled vesicles is shown in Figure 1d3.

Figure 1. Formation of SSLBs and SSLB array assembly. (a) Illustration of a SSLB composed of 3 types of lipids on an SiO2 bead: egg PC (black), Rho-DPPE (red) and GM1 (blue). (b1-d1) Process flow for assembly of a SSLB array showing SSLBs scattered on a microwell array (b1), use of a PDMS squeegee to clear the surface of SSLBs not settled into microwells (c1), and the final SSLB array (d1). (b2-d2) The same process illustrated for the case of phospholipid vesicles or natural membrane particles where more than one vesicle or particle can be accommodated in each microwell. (b3) A fluorescence image of phospholipid vesicles adsorbed onto a microwell substrate corresponding to the illustration in (b2). (d3) A fluorescence image of a microarray composed of phospholipid vesicles corresponding to the illustration in (d2). Adapted from references 20 and 21 and used with permission of the American Chemical Society.
Individual SiO2 beads supports for SSLBs in microwells were imaged with scanning electron microscopy (SEM) from both the angled top-view and cross-sectional perspectives. A top view of an area approximately 15 µm x 15 µm is shown in Figure 2a, while Figure 2b shows a cross-sectional view of a single bead immobilized in a microwell. The lighter layer coating the microwell is 100 nm of Al2O3 deposited by atomic layer deposition to tune the well diameter so that the wells are capable of accommodating only single beads.

Figure 2. Scanning electron microscopy images of SSLB arrays. (a) An electron micrograph of a 15 mm x 15 mm area showing SSLB bead supports immobilized in microwell arrays. (b) A single 700 nm diameter SSLB in a microwell. The microwell coating is Al2O3, which is deposited to shrink the microwell so that only individual beads can fit inside. Adapted from reference 20 and used with permission of the American Chemical Society.
Once the SSLB arrays are prepared, they can be imaged by fluorescence microscopy. Figure 3a shows a 100 µm × 100 µm area of an array with 936 SSLBs. To determine the percent of occupancy of an array, the fluorescence image can be compared with a brightfield image of the same area. Figure 3b shows an area with 550 microwells and 416 SSLBs for an occupancy rate of 76%.
Sequential deposition of different types of SSLBs was used to create arrays with more than one type of SSLB, where the identity of individual SSLBs was determined by the absence or presence of a toxin receptor (GM1) and its binding of cholera toxin (CTx). The assembly process was the same as for a single SSLB type but was carried out a second time with a different SSLB identity. The starting concentration of SSLBs was 10x lower to reduce occupancy of the first type of SSLB and to allow more vacancies to be occupied by the second type of SSLB. In the Figures 3c-3e, an SSLB array with two types of SSLBs is shown. All of the SSLBs contain red fluorescent Rho-DPPE (Figure 3c), but only some of the SSLBs contain GM1, a ganglioside that is the receptor for the B-subunit of CTx. When exposed to green fluorescent Alexa 488-conjugated CTx, only the SSLBs containing GM1 fluoresce in the green channel (Figure 3d). When the red and green images are merged, it is possible to determine which SSLBs contain GM1 (yellow) and which do not (red) (Figure 3e).

Figure 3. Fluorescence imaging of SSLB arrays. (a) A 100 µm x 100 µm area containing 936 Rho-DPPE-labeled egg PC SSLBs. (b) A brightfield and fluorescence overlay showing a SSLB with 76% of the microwells occupied. (c-e) A SSLB array containing SSLBs that contain Rho-DPPE (c), a fraction of which also contain GM1, which is bound by Alexa-488-conjugated CTx (d). (e) Merger of red and green channels where colocalized fluorescence indicates the presence of GM1 on the SSLB. The blue circles indicate SSLBs that only fluoresce red, i.e. lacking GM1. Adapted from reference 20 and used with permission of the American Chemical Society.
Fluorescence data from SSLB arrays was analyzed by measuring the intensity of individual SSLBs in an image. The SSLB intensity data from arrays were compiled into histograms. Figure 4a shows one such histogram from the analysis of Rho-DPPE fluorescence from a SSLB array composed solely of SSLBs containing egg PC and 1 mol% Rho-DPPE. The data in Figure 4a were acquired by analyzing the image in Figure 3a. We found the SSLB arrays to be robust and stable for at least one week when refrigerated and submerged in buffer. The arrays did not lose any fluorescence intensity, nor did the occupancy of the arrays change over the course of one week (Figure 4b).

Figure 4. (a) Histogram showing the distribution of fluorescence intensities for the SSLB array shown in Figure 3a. The mean intensity for each SSLB in the array was used to compile the histogram. The solid line is a Gaussian fit to the data. (b) Mean array occupancy (black circles) and fluorescence intensity (red squares) for a SSLB array monitored over the course of one week. Adapted from reference 20 and used with permission of the American Chemical Society.
We made arrays composed of SSLBs with varying concentrations of GM1 (0, 0.5, 1 and 2 mol%) and exposed them to a fixed concentration of CTx as well as arrays with SSLBs with concentrations of GM1 and exposed them to varying concentrations of CTx. The later experiment was used to determine the equilibrium dissociation constant (Kd) for GM1/CTx binding. The SSLB arrays with varying GM1 concentration were exposed to 50 nM Alexa-488 labeled CTx for 1 hr then washed and imaged. The intensity histograms for SSLB arrays with 0.5 – 2 mol% GM1 exposed to 50 nM Alexa 488-conjugated CTx can be seen in Figure 5a. Figure 5b shows the linear dependence of mean fluorescence intensity upon GM1 concentration. When the concentration of GM1 in SSLBs is fixed at 2 mol% and the SSLB arrays are exposed to CTx ranging from 16 pM to 158 nM, the binding response follows sigmoidal curves as shown in Figure 5c. The curves fit to this data are based upon two different binding models: the Langmuir isotherm (Eq. 1) and the Hill-Waud model (Eq. 2). Please click here to view a larger version of this figure.
 (1)
(1)
 (2)
(2)
Where F is the fluorescence intensity at a given CTx concentration, Fmax is the fluorescence intensity when binding is saturated, [CTx] is the CTx concentration, KD and KH are the dissociation constants from the Langmuir and Hill-Waud fits, respectively, and n is cooperativity coefficient in the Hill-Waud model. The cooperativity coefficient (n) estimates the amount of multivalent binding, where an n greater than one indicates that each CTx molecule binds more than one GM1. The concentration of CTx at 0.5 x Fmax is the KD or KH for the GM1/CTx interaction. In our experiments we calculated a KD of 1.6 ± 0.2 nM and KH of 1.4 ± 0.2 nM with an n value of 1.3, indicating cooperative binding. The dissociation constants for the GM1/CTx interaction vary widely in the literature, ranging over 5 orders of magnitude from 4.5 pM to 370 nM26,27.
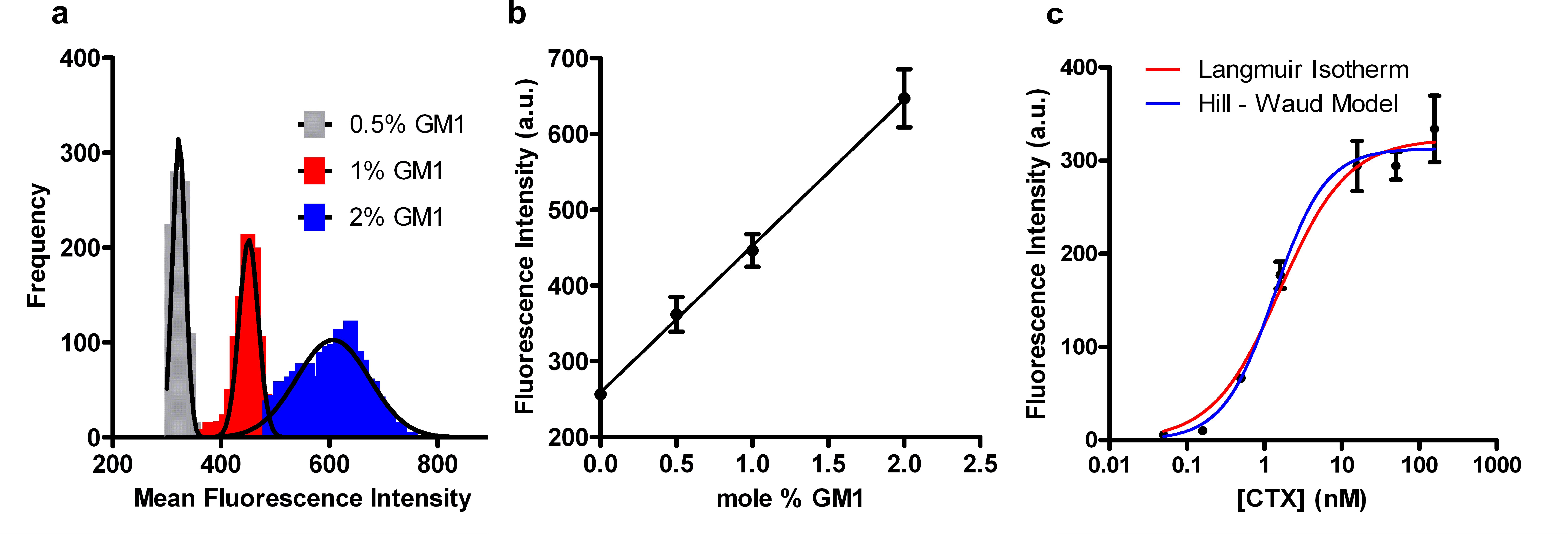
Figure 5. CTx/GM1 binding assays on SSLB arrays. (a) Histograms of the fluorescence intensity of individual SSLBs within SSLB arrays. The arrays contained SSLBs with 0.5, 1 or 2 mol% GM1 and were exposed to 50 nM Alexa 488-conjugated CTx. The solid lines are Gaussian fits. (b) Plot of the distribution means from (a) as a function of GM1 concentration. (c) Binding curves showing the distribution means from arrays containing SSLBs with 2 mol% GM1 after exposure to Alexa-488-conjugated CTx varying from 16 pM to 158 nM. The lines are fits to the Langmuir isotherm (red) and Hill-Waud (blue) binding models. The error bars in (b-c) represent the standard deviations of the distributions of fluorescence intensity corresponding to each data point. Adapted from reference 20 and used with permission of the American Chemical Society.
As mentioned earlier, it is also possible to form arrays with biomembrane material derived from natural sources21. The arrays are formed the same way, by dispersing membrane particles on a microwell substrate, then cleaning the top surface with the squeegee to leave behind membrane particles in the microwells. Figure 6 shows examples of myelin and neuronal lipid raft microarrays. In Figure 6a, myelin particles are labeled with the lipophilic fluorophore FM1-43. Lipid rafts are known to be enriched in cholesterol and gangliosides such as GM1; thus, Alexa 488-conjugated CTx binds strongly to lipid raft microarrays as shown in Figure 6b, while fluorescently labeled streptavidin (SAPE) does not bind to the array. (Figure 6c) We also created stripe arrays by delivering myelin and lipid raft particles to the microwell substrate with a microfluidic chip with 250 µm-wide channels. After particle delivery, the microfluidic chip was detached from the microwell substrate and the squeegee process carried out as usual. The stripe arrays were then exposed to a fluorescent anti-oligodendrocyte IgM antibody (IgM O4), which binds sulfatide found in myelin. (Figure 7) The myelin and lipid raft stripe microarrays in Figure 7 are shown at increasing levels of magnification, and at the highest level of magnification, it is readily apparent that IgM O4 only binds to the myelin microarrays because sulfatide is absent from the lipid rafts.

Figure 6. Myelin particle and neuronal lipid raft particle arrays. (a) A myelin particle microarray where the myelin particles are rendered fluorescent by the lipophilic fluorophore FM1-43. The dashed line indicates the edge of the array area. (b) Lipid raft microarray showing CTx binding to the lipid raft particles. (c) Lipid raft microarray exposed to fluorescent streptavidin-phycoerythrin (SAPE) showing little to no nonspecific binding. Adapted from reference 21 and used with permission of the American Chemical Society.

Figure 7. Multicomponent arrays formed by microfluidic delivery of myelin and neuronal raft membranes. (a) Fluorescence image after myelin and rafts were deposited via microfluidic channels on the microarray substrate. The left and right stripes contain myelin and the middle stripe contains neuronal raft membranes. The membranes are stained with FM1-43, which labels all lipid material. The scale bar is 250 μm. (b) Fluorescence image of the same three stripes after applying the PDMS squeegee and incubating with IgM O4. The scale bar is 250 μm. (c-e) Magnified images from the three stripes showing that IgM O4 only binds to the myelin microarrays: (c) left stripe and (e) right stripe. The scale bars in panels c-e are 30 μm. Adapted from reference 21 and used with permission of the American Chemical Society.
Discussion
In this work we show that monodisperse SiO2 beads coated with supported lipid bilayers can be arrayed into microwell arrays without the need for targeting ligands on the lipid bilayers or the substrate surface, and the arrays can be used for characterizing toxin-lipid interactions. The dissociation constant we calculated for CTx/GM1 binding compares favorably, given the wide disparity of values in the literature, with a previous report by Winter et al., where colloidal assembly of lipid-coated beads was used to calculate a dissociation constant of about 30 nM. By fitting the data to the Hill-Waud binding model, we determined that the CTx/GM1 interaction is likely multivalent, which agrees with previous reports28. The B (ganglioside binding) subunit of CTx exists as a pentamer, with each of the five units capable of binding a GM1 molecule29; thus, it is not surprising that more than one subunit may be bound given the diffusive mobility of GM1 in SLBs30.
Arrays of chemically functionalized beads have been used in other analytical studies31. In these types of experiments, beads are typically immobilized in microwells created in the end of etched optical fibers. Multiplex fluorescence assays can be carried out in this configuration by mixing subpopulations of beads and immobilizing them simultaneously onto the end of the optical fiber32. In our multiplex assay (Figures 3c-3e), we immobilized optically encoded SSLBs sequentially because lipids on SSLBs free in solution could be transferred from one type of SSLB to another33. While this adds extra steps in the preparation of SSLB arrays, it not overly time consuming and is the most efficient approach for lipid-functionalized beads. We have also spatially encoded arrays by depositing different SSLB populations in adjacent microwell arrays, and there is no observable cross-talk between the arrays20. To reduce sample-preparation times, SSLB arrays can be prepared in advance and used when desired. We tested the stability of arrays over the course of one week and found no significant degradation of the arrays in terms of fluorescence intensity, which indicates that the amount of lipid material on the SSLBs does not change with time. Also, the array occupancy does not change over time, which suggests that once SSLBs are immobilized, they do not detach from the microwells (Figure 4b).
In addition to beads coated with synthetic lipid bilayers, we have also used this method to create arrays of natural membrane particles21. In that work, we immobilized myelin particles derived from mouse brain as well as neuronal membrane particles, such as the lipid-raft fraction. We were able to use the arrays to identify cell-specific membrane surface molecules by antibody binding (Figures 6 and 7). Furthermore, when the size of the microwells was reduced to the nanoscale and the top surface of the array was coated with gold, we were able to employ surface plasmon resonance to monitor binding of antibodies to myelin membrane particles without the use of fluorescent labels. The major difference between using SSLB and vesicles or natural membrane particles to form arrays is that in the case of SSLBs the exact amount of membrane material in each microwell is known because only a single bead can fit in each well. With vesicles or natural membrane particles, there is no way to control the amount of material in each microwell, other than to adjust the starting concentration of vesicles or particles. However, the well-to-well variability of the number of vesicles is not very large compared to the distribution of vesicle diameters21.
Finally, this method can have diverse future applications. Junesch and coworkers have employed a squeegee to remove nanoparticles during colloidal lithography nanofabrication34. Membrane arrays could be formed to mimic cell-cell contacts to investigate cellular focal adhesion35 or examine the binding of membrane curvature-sensing proteins36. In addition, the use of porous beads would facilitate the inclusion of transmembrane proteins into the lipid bilayers37 or local delivery of stored cargo to cells cultured on a biomembrane array substrate. The microwell platform is fabricated by photolithography, which allows for design flexibility. Here our arrays had hexagonal geometry, but square, linear, or other geometries could be created with varying array periodicity and microwell dimensions to investigate the spatial effects of focal adhesion.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by grants to S.H.O. from the National Institutes of Health (R01 GM092993), the National Science Foundation (NSF CAREER Award and DBI 0964216), the Office of Naval Research (ONR) Young Investigator Program and the Minnesota Partnership Award for Biotechnology and Medical Genomics. Device fabrication was performed at the University of Minnesota Nanofabrication Center (NFC), which receives support from the NSF through the National Nanotechnology Infrastructure Network. This work was also supported by grants to M.R. from the National Institutes of Health (NS048357, R21 NS073684), the National Multiple Sclerosis Society (CA1060A11), the Applebaum, Hilton, Peterson and Sanford Foundations and the McNeilus family. The authors wish to thank Hyungsoon Im for assistance with illustrations and Shailabh Kumar for assistance with scanning electron microscopy.
Materials
| 4-inch silicon wafers | University Wafer | 425 | |
| Shipley MEGAPOSIT SPR955-CM 0.7 photoresist | MicroChem | SPR955-CM | |
| Shipley MICROPOSIT CD-26 developer | MicroChem | CD-26 | |
| i-line stepper | Canon | 2500 i3 stepper | |
| Vision 320 reactive ion etcher | Advanced Vacuum | Vision 320 RIE | |
| Deep trench reactive ion etcher | Plasma Therm | SLR-770 | |
| Atomic layer depostion system | Cambridge NanoTech | Savannah | |
| Dow Corning Sylgard 184 poly(dimethylsiloxane) kit | Ellsworth Adhesives | 184 SIL ELAST KIT 0.5KG | |
| egg phosphatidylcholine | Avanti Polar Lipids | 840051C | |
| 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) ammonium salt | Avanti Polar Lipids | 810158C | |
| monosialoganglioside GM1 | Avanti Polar Lipids | 860065P | |
| Silica beads | Bangs Laboratories | SS03N/4666 | Packaging on the bead container states the beads are 900 nm in diameter. However, after light-scattering and electron microscopy we determined the beads are roughly 700 nm in diameter. |
| Cholera toxin B-subunit, Alexa 488 conjugate | Molecular Probes | C-34775 | |
| Anti-oligodentrocyte antibody IgM O4, NorthernLights 557 conjugate | R&D Systems | NL1326R | |
| FM1-43 | Molecular Probes | T-3136 | |
| Eppendorf MiniSpin centrifuge | Fisher Scientific | 05-401-09 |
References
- Drews, J. Drug discovery: A Historical Perspective. Science. 287 (5460), 1960-1964 (1126).
- Cooper, M. A. Advances in Membrane Receptor Screening and Analysis. J. Mol. Recognit. 17 (4), 286-315 (2004).
- Voskuhl, J., Ravoo, B. J. Molecular Recognition of Bilayer Besicles. Chem. Soc. Rev. 38 (2), 495-505 (2009).
- Walde, P., Cosentino, K., Engel, H., Stano, P. Giant Vesicles: Preparations and Applications. Chembiochem. 11 (7), 848-865 (1002).
- Castellana, E. T., Cremer, P. S. Solid Supported Lipid Bilayers: From Biophysical Studies to Sensor Design. Surf. Sci. Rep. 61 (10), 429-444 (2006).
- Johnson, J. M., Ha, T., Chu, S., Boxer, S. G. Early Steps of Supported Bilayer Formation Probed by Single Vesicle Fluorescence Assays. Biophys. J. 83 (6), 3371-3379 (2002).
- Krysinski, P., Zebrowska, A., Michota, A., Bukowska, J., Becucci, L., Moncelli, M. R. Tethered Mono- and Bilayer Lipid Membranes on Au and Hg. Langmuir. 17 (13), 3852-3857 (2001).
- Li, L., Wang, H. F., Cheng, J. X. Quantitative Coherent anti-Stokes Raman Scattering Imaging of Lipid Distribution in Coexisting Domains. Biophys. J. 89 (5), 3480-3490 (2005).
- Richter, R. P., Brisson, A. R. Following the Formation of Supported Lipid Bilayers on Mica: A Study Combining AFM, QCM-D, and Ellipsometry. Biophys. J. 88 (5), 3422-3433 (2005).
- Dahlin, A., Zäch, M., Rindzevicius, T., Käll, M., Sutherland, D. S., Höök, F. Localized Surface Plasmon Resonance Sensing of Lipid-Membrane-Mediated Biorecognition Events. J. Am. Chem. Soc. 127 (14), 5043-5048 (2005).
- Kraft, M. L., Weber, P. K., Longo, M. L., Hutcheon, I. D., Boxer, S. G. Phase Separation of Lipid Membranes Analyzed with High-Resolution Secondary Ion Mass Spectrometry. Science. 313 (5795), 1948-1951 (2006).
- Groves, J. T., Boxer, S. G. Micropattern Formation in Supported Lipid Membranes. Acc. Chem. Res. 35 (3), 149-157 (2002).
- Bally, M., Bailey, K., Sugihara, K., Grieshaber, D., Vörös, J., Stadler, B. Liposome and Lipid Bilayer Arrays Towards Biosensing Applications. Small. 6 (22), 2481-2497 (2010).
- Groves, J. T., Dustin, M. L. Supported Planar Bilayers in Studies on Immune Cell Adhesion and Communication. J. Immunol. Methods. 278 (1-2), 19-32 .
- Yang, T. L., Jung, S. Y., Mao, H. B., Cremer, P. S. Fabrication of Phospholipid Bilayer-Coated Microchannels for On-Chip Immunoassays. Anal. Chem. 73 (2), 165-169 (2001).
- Groves, J. T., Ulman, N., Boxer, S. G. Micropatterning Fluid Lipid Bilayers on Solid Supports. Science. 275 (5300), 651-653 (1997).
- Hovis, J. S., Boxer, S. G. Patterning and Composition Arrays of Supported Lipid Bilayers by Microcontact Printing. Langmuir. 17 (11), 3400-3405 (2001).
- Yee, C. K., Amweg, M. L., Parikh, A. N. Direct Photochemical Patterning and Refunctionalization of Supported Phospholipid Bilayers. J. Am. Chem. Soc. 126 (43), 13962-13972 (2004).
- Kelly, C. V., Craighead, H. G. Nanofabrication for the Analysis and Manipulation of Membranes. Ann. Biomed. Eng. 40 (6), 1356-1366 (2012).
- Wittenberg, N. J., Johnson, T. W., Oh, S. H. High-Density Arrays of Submicron Spherical Supported Lipid Bilayers. Anal. Chem. 84 (19), 8207-8213 (2012).
- Wittenberg, N. J., et al. Facile Assembly of Micro- and Nanoarrays for Sensing with Natural Cell Membranes. ACS Nano. 5 (9), 7555-7564 (2011).
- Stamou, D., Duschl, C., Delamarche, E., Vogel, H. Self-Assembled Microarrays of Attoliter Molecular Vessels. Angew. Chem. Int. Ed. 42 (45), 5580-5583 (2003).
- Kalyankar, N. D., et al. Arraying of Intact Liposomes into Chemically Functionalized Microwells. Langmuir. 22 (12), 5403-5411 (2006).
- Dahlin, A. B., Jonsson, M. P., Höök, F. Specific Self-Assembly of Single Lipid Vesicles in Nanoplasmonic Apertures in Gold. Adv. Mater. 20 (8), 1436-1442 (2008).
- Nair, P. M., Salaita, K., Petit, R. S., Groves, J. T. Using Patterned Supported Lipid Membranes to Investigate the Role of Receptor Organization in Intercellular Signaling. Nat. Protoc. 6 (4), 523-539 (2011).
- Kuziemko, G. M., Stroh, M., Stevens, R. C. Cholera Toxin Binding Affinity and Specificity for Gangliosides Determined by Surface Plasmon Resonance. Biochemistry. 35 (20), 6375-6384 (1996).
- Moran-Mirabal, J. M., Edel, J. B., Meyer, G. D., Throckmorton, D., Singh, A. K., Craighead, H. G. Micrometer-Sized Supported Lipid Bilayer Arrays for Bacterial Toxin Binding Studies Through Total Internal Reflection Fluorescence Microscopy. Biophys. J. 89 (1), 296-305 (2005).
- Shi, J. J., Yang, T. L., Kataoka, S., Zhang, Y. J., Diaz, A. J., Cremer, P. S. GM1 Clustering Inhibits Cholera Toxin Binding in Supported Phospholipid Membranes. J. Am. Chem. Soc. 129 (18), 5954-5961 (2007).
- Gill, D. M. The Arrangement of Subunits in Cholera Toxin. Biochemistry. 15 (6), 1242-1248 (1021).
- Weng, K. C., Kanter, J. L., Robinson, W. H., Frank, C. W. Fluid Supported Lipid Bilayers Containing Monosialoganglioside GM1: A QCM-D and FRAP study. Colloid Surface B. 50 (1), 76-84 (1016).
- Walt, D. R. Fibre Optic Microarrays. Chem. Soc. Rev. 39 (1), 38-50 (2010).
- Bake, K. D., Walt, D. R. Multiplexed Spectroscopic Detections. Annu. Rev. Anal. Chem. 1 (1), 515-547 (2008).
- Kundu, J., Levin, C. S., Halas, N. J. Real-Time Monitoring of Lipid Transfer Between Vesicles and Hybrid Bilayers on Au Nanoshells Using Surface Enhanced Raman Scattering (SERS). Nanoscale. 1 (1), 114-117 (2009).
- Junesch, J., Sannomiya, T., Dahlin, A. B. Optical Properties of Nanohole Arrays in Metal-Dielectric Double Films Prepared by Mask-on-Metal Colloidal Lithography. ACS Nano. 6 (11), 10405-10415 (2012).
- Wu, M., Holowka, D., Craighead, H. G., Baird, B. Visualization of Plasma Membrane Compartmentalization with Patterned Lipid Bilayers. Proc. Natl. Acad. Sci. U.S.A. 101 (38), 13798-13803 (2004).
- Hatzakis, N. S., Bhatia, V. K., Larsen, J., Madsen, K. L., Bolinger, P. Y., Kunding, A. H., Castillo, J., Gether, U., Hedegard, P., Stamou, D. How Curved Membranes Recruit Amphipathic Helices and Protein Anchoring Motifs. Nat. Chem. Biol. 5 (11), 835-841 (2009).
- Roizard, S., Danelon, C., Hassaine, G., Piguett, J., Schulze, K., Hovius, R., Tampe, R., Vogel, H. Activation of G-Protein-Coupled Receptors in Cell-Derived Plasma Membranes Supported on Porous Beads. J. Am. Chem. Soc. 133 (42), 16868-16874 (2011).

